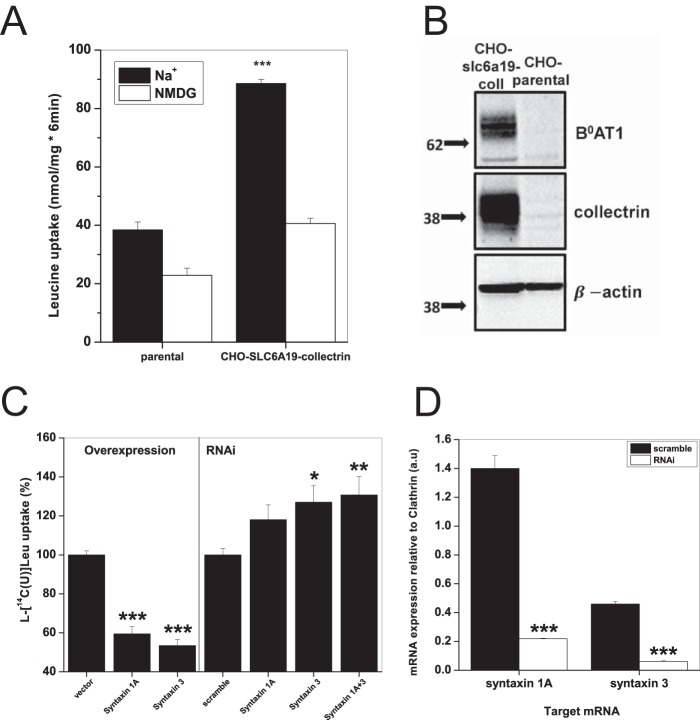
FIGURE 7.
Syntaxin 1A and syntaxin 3 inhibit B0AT1 activity. A, uptake of 100 μm l-[U-14C]leucine was measured over 6 min in CHO parental and CHO-SLC6A19-collectrin cells in either Hanks' buffered saline (Na+) or with NMDG replacing sodium (NMDG). Each bar represents mean transport activity ± S.D. (e = 3, n = 3, ***, p = 0.005 level). p values are calculated as difference from Na+-independent uptake condition. B, immunodetection of B0AT1 and collectrin in whole membrane preparations of CHO parental and CHO-SLC6A19-collectrin cells; 20 μg of total protein was loaded into each well. Molecular mass markers (in kDa) are indicated on the left, and detected proteins are indicated on the right. C, CHO-SLC6A19-collectrin cells were transfected with 3 μg of pcDNA3.1+ vector DNA encoding the indicated murine syntaxin gene (overexpression, left panel) or 50 pmol of siRNA against the CHO endogenous syntaxins (RNAi, right panel). 100 μm l-[U-14C]leucine uptake in CHO-SLC6A19-collectrin cells was measured over 6 min in either Hanks' buffered saline (Na+) or with NMDG replacing sodium (NMDG). The ratio of net sodium-dependent/sodium-independent uptake for each uptake condition was normalized to the pcDNA3.1+ vector-only control (overexpression) or scrambled siRNA (RNAi). Each bar represents mean ± S.E. (n = 3, e = 3, ***, p 0.005 level; **, p 0.01 level; *, p 0.05 level). p values are calculated as difference from control conditions, vector only (overexpression) or scramble (RNAi). D, levels of syntaxin 1A and syntaxin 3 mRNA transcripts were estimated by RT-PCR with 25 cycles of amplification. Closed bars represent control samples using scrambled RNAi, and open bars show the effect of specific RNAi. Samples were run on agarose gel, and quantification was made relative to clathrin mRNA expression in the same sample. Each bar represents mean ± S.E. (e = 3, ***, p 0.005 level). p values are calculated as difference from scramble conditions.