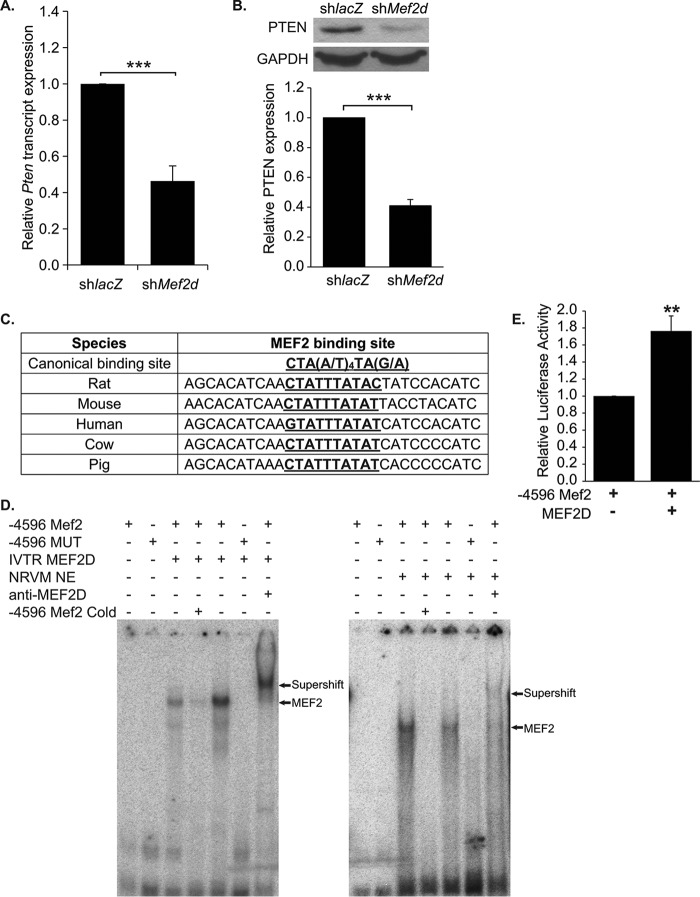
FIGURE 4.
PTEN expression is down-regulated in MEF2D deficient NRVMs. A, Pten transcripts were down-regulated 2.5-fold in cardiomyocytes depleted of MEF2D. B, Western blot analysis and densitometry confirm decreased levels of PTEN expression, with a 2.5-fold reduction of PTEN protein. C, the MEF2 binding site and flanking sequence located 4596 bases upstream of the putative Pten transcriptional start site is highly conserved between species. D, gel shift assay reveals binding of both in vitro translated and endogenous MEF2D to the wild-type (4596 Mef2) but not mutant (−4596 MUT) Mef2 site identified within the rat Pten regulatory region. Incubation with MEF2D antibodies shift the MEF2 complex bound to the radiolabeled −4596 Mef2 sequence. Binding of MEF2D to the −4596 Mef2 site is competed by the unlabeled wild-type but not the mutant sequence. E, luciferase analysis of the wild-type PTEN promoter fragment containing an evolutionarily conserved MEF2 binding site (−4596 Mef2). HEK293T cells were transfected with mouse MEF2D (n = 5). The data are means ± S.E. (error bars). **, p < 0.01; ***, p < 0.001.