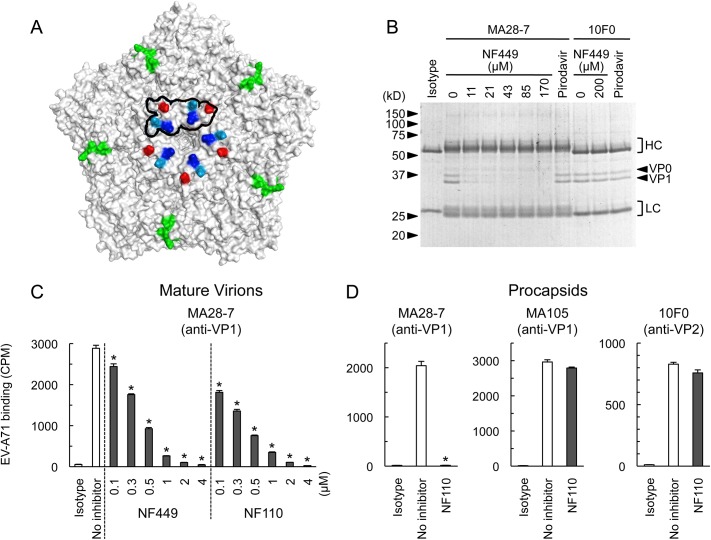
Fig 9. NF449 and NF110 specifically prevent attachment of a monoclonal antibody to the viral 5-fold vertex.
(A) An EV-A71 pentamer shown with the 5-fold vertex at the center. VP1-98 (red) was mutated in an NF449 escape mutant. VP1-242K (light blue) is involved in virus interaction with PSGL-1. VP1-244K (dark blue) is implicated both in virus interactions with both PSGL-1 and NF449. The MA28-7 footprint, as determined by cryo-electronmicroscopy, is outlined in black. The VP2 epitope recognized by mAb 10F0 is indicated in green. (B) NF449 inhibits immunoprecipitation of EV-A71-1095 by MA28-7 but not by 10F0. EV-A71-1095 concentrated from supernatants of infected cells was incubated with MA28-7 or 10F0 fixed to Protein G beads, in the presence of NF449 at the indicated concentrations, or 200 μM Pirodavir; beads were washed, and immunoprecipitated proteins were examined on Coomassie-stained gels. Molecular weight markers are shown at the left. Arrows at the right indicate capsid proteins VP0 and VP1, and brackets indicate antibody heavy (HC) and light chains (LC). (C) NF449 and NF110 inhibit attachment of MA28-7 to mature virions. Purified 35S-labeled mature virions were incubated with MA28-7-coated beads, in the presence of inhibitors as indicated. (D) NF110 inhibits attachment of MA28-7, but not 10F0 or MA105, to procapsids. Purified 35S-labeled procapsids were incubated with monoclonal antibodies fixed to Protein G beads, in the presence of 20 μM NF110. Results are indicated as the mean and S.D. for triplicate samples. Asterisks indicate P < 0.01 compared to the no inhibitor control.