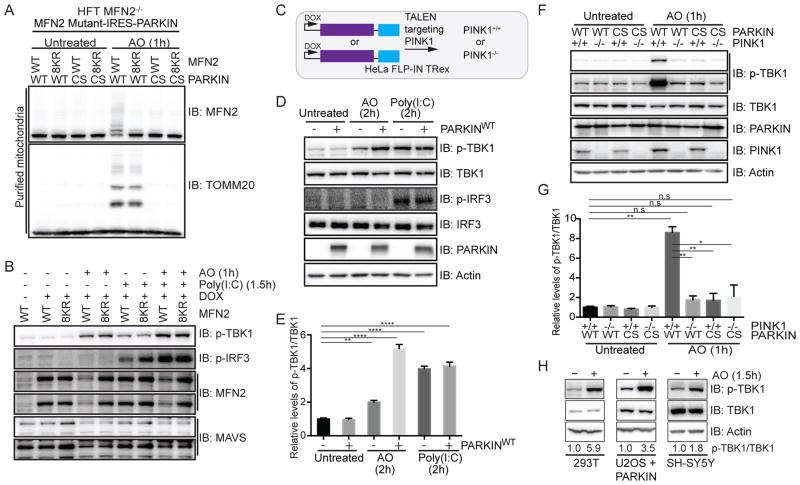
Figure 1. Activation of TBK1 upon mitochondrial depolarization requires PARKIN and PINK1.
(A,B) HFT-MFN2−/− cells expressing an inducible MFN2 (WT or 8KR mutant)-IRES-PARKIN (WT or CS mutant) were depolarized (1h) and purified mitochondria (20μg) subjected to immunoblotting with MFN2 or TOMM20 antibodies (Panel A). In parallel experiments, cells were either left untreated, exposed to AO (1h) or transfected with Poly(I:C) (10μg/ml, 1.5h) and lysated subjected to immunoblotting with the indicated antibodies (Panel B).
(C) Schematic of cell system used in this work. HeLa Flp-in T-Rex (HFT) cells harboring PARKINWT or catalytically inactive PARKINCS were created in the context of PINK1+/+ or PINK1−/− generated using gene-editing with a TALEN targeting PINK1.
(D,E) HFT or HFT-PARKINWT cells were treated with AO (2h) to depolarize mitochondria or Poly(I:C) (2h) to activate the RIG-I/MDA5/MAVS pathway. Lysates were subjected to immunoblotting with the indicated antibodies (Panel D). Quantification of relative p-TBK1S172/TBK1 levels for biological triplicate experiments (Panel E). Error bars represent SEM from triplicate experiments.
(F,G) HFT cells with the indicated PARKIN and PINK1 genotypes were depolarized with AO (1h) and lysates subjected to immunoblotting with the indicated antibodies (Panel F). Quantification of relative p-TBK1/TBK1 levels for biological triplicate experiments (Panel G). Error bars represent SEM from triplicate experiments.
(H) U2OS cells stably expressing PARKINWT or SH-SY5Y cells were left untreated or depolarized with AO (1h) prior to immunoblotting of lysates. The p-TBK1S172/TBK1 ratios are indicated.
Panels E and G analyzed by one-way ANOVA with Dunnett’s multiple comparisons test (n=3 biological replicates). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. n.s., not significant. See also Figure S1.