Fig. 1.
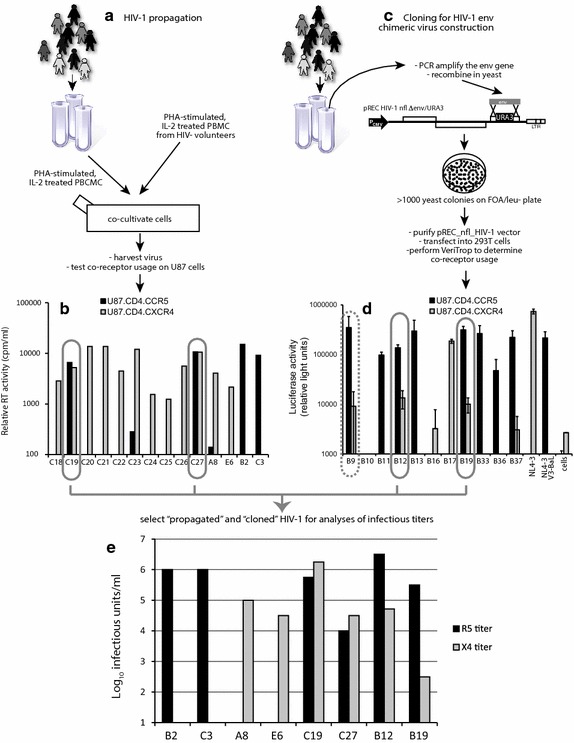
Identification of dual/mixed isolates following HIV-1 propagation or construction of env chimeric viruses. a HIV-1 subtype C isolates from Zimbabwe were propagated by PBMC co-cultivation as described [46] and in the “Methods”. c The HIV-1 env coding region of HIV-1 subtype B infected patients was PCR amplified and then cloned into pREC_nfl_HIV-1nl4-3Δgp120/URA3 via yeast recombination/gap repair [47]. The propagated HIV-1 isolates (a) and cloned env genes expressed following 293T transfection (c) were screened for co-receptor usage by infection(1) (b) or cell-to-cell fusion (d) (Veritrop; [48]) on U87.CD4 cells expressing either CXCR4 or CCR5(2). In the virus infection system (b), virus production was monitored by RT activity in supernatant [87]. In the cell-to-cell fusion assay (d), the ability of pREC_nfl_envptX to modulate receptor binding and cell fusion was monitored by firefly luciferase activity, i.e. Rev/Tat in the effector cell controlling Luc expression under the control of the HIV-1 LTR and rescued via the RRE housed in an intron with Luc [80]. Two primary HIV-1 isolates and two env chimeric viruses were propagated to measure infectious titers on PBMCs (Additional file 1: Table S1), U87.CD4.CCR5 and U87.CD4.CXCR4 cells (e) using the classical Reed–Munch approach [79].
