Figure 2.
Link between chromothripsis and hyperploidy
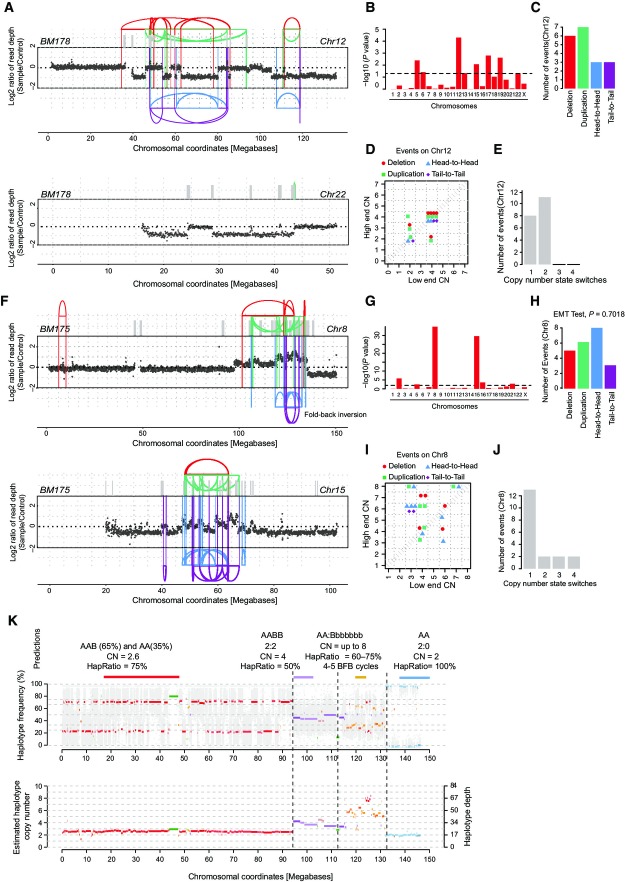
- DNA alteration patterns of chromosomes 12 and 22 in BM178 based on mate-pair and WGS data, with highly oscillating copy number profiles consistent with the occurrence of chromothripsis. SRs are color-coded: red, deletion type (T-H); green, duplication type (H-T); blue, head-to-head (H-H) type; purple, tail-to-tail (T-T) type; gray, inter-chromosomal.
- Statistically significant deviation from null hypothesis of no breakpoint clustering in BM178 (y-axis depicts -log10(P-value) for KS test applied to each chromosome), in line with the occurrence of chromothripsis (see Appendix notes, inference of chromothripsis, criterion 1).
- Randomness of DNA fragment joins (Korbel & Campbell, 2013) for BM178 chromosome 12 (EMT test, P = 0.5041), additionally supporting the occurrence of chromothripsis. P-value derived from multinomial testing against null hypothesis “equal distribution of joins” (see Appendix notes, inference of chromothripsis, criterion 2).
- Copy number jump distribution (Li et al, 2014) for chromosome 12 in BM178. Points on the diagonal indicate that chromothripsis occurred on a previously intact chromosome. (Appendix notes, inference of chromothripsis, criterion 3).
- Copy number segment switches (Li et al, 2014) for chromosome 12 in BM178, consistent with chromothripsis occurring as the initial event (see Appendix notes, inference of chromothripsis, criterion 3).
- DNA alteration patterns of chromosomes 8 and 15 in BM175 based on mate-pair and high-coverage WGS data, with highly oscillating copy number profiles consistent with the occurrence of chromothripsis.
- Statistically significant deviation from null hypothesis of no breakpoint clustering in BM175 (y-axis depicts -log10(P-value) for KS test applied to each chromosome), in line with the occurrence of chromothripsis.
- Randomness of DNA fragment joins (Korbel & Campbell, 2013) for BM175 chromosome 8 (EMT test, P = 0.7018), additionally supporting the occurrence of chromothripsis.
- Copy number jump distribution (Li et al, 2014) between joined fragments of chromosome 8 in BM175 (displayed for segments with confident copy number ascertainment). Most copy number jumps are off the diagonal, indicating that chromothripsis occurred on a previously rearranged chromosome.
- Copy number segment switches (Li et al, 2014) for chromosome 8 in BM175, consistent with BFBs occurring prior to chromothripsis.
- Evidence for chromothripsis occurring on one single parental haplotype. Based on the haplotype frequencies, and the copy number estimates, we inferred that the majority of the chromosome 8 is present in 2 copies, but possibly due to an early subclonal event, two subclones exhibiting separate haplotype ratios (AA and AAB) exist. The shift in the haplotype ratio and the increase in copy number suggest that the B haplotype underwent several (up to 5) cycles of BFB as well as a chromothripsis event, resulting in amplified segments of as high as 8 copy numbers. The sharp drop close to the chromosome end is associated with a shift to a haplotype ratio of 0, that is, complete loss of the B haplotype (in line with the BFB and chromothripsis events occurring on the same haplotype). Predictions of haplotype segments are indicated in the above panel.