Figure 3.
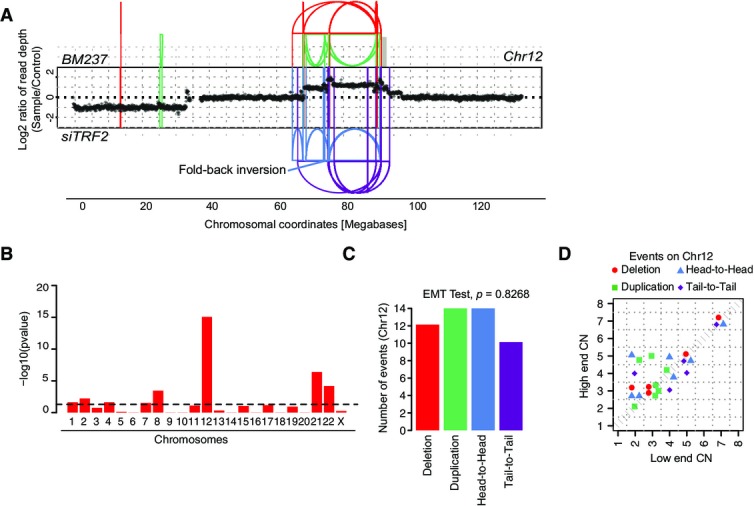
Evidence for chromothripsis in TRF2-depleted cells
- DNA alteration patterns of chromosome 12 in BM237 based on mate-pair data. Highly oscillating copy number profiles are consistent with the occurrence of chromothripsis. SRs are color-coded: red, deletion type (T-H); green, duplication type (H-T); blue, head-to-head (H-H) type; purple, tail-to-tail (T-T) type; gray, inter-chromosomal.
- Statistically significant deviation from null hypothesis of no breakpoint clustering in BM237 (y-axis depicts -log10(P-value) for KS test applied to each chromosome), in line with the occurrence of chromothripsis.
- Randomness of DNA fragment joins for BM237, additionally supporting the occurrence of chromothripsis. P-value is derived from multinomial testing against null hypothesis “equal distribution of joins.”
- Copy number jump distribution between joined fragments of chromosomes undergoing chromothripsis (displayed for segments with confident copy number ascertainment). On chromosome 12 in BM237, chromothripsis occurred most likely in conjunction with breakage–fusion–bridge cycles.
