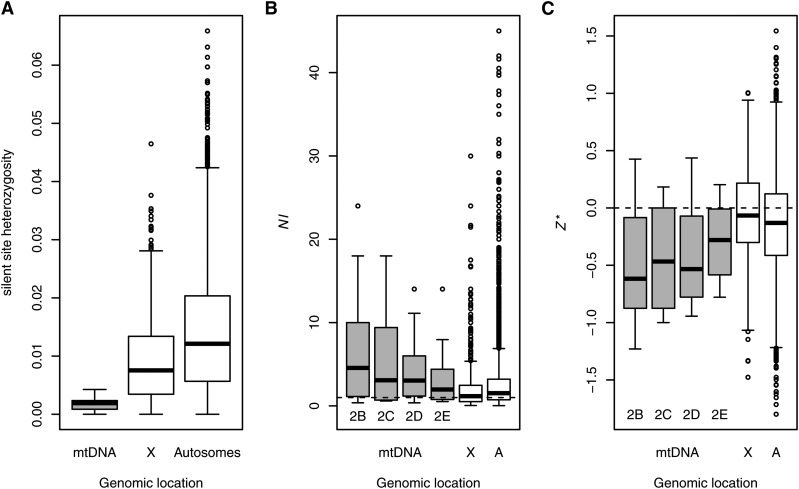
Figure 1.
Effect of genomic location on silent-site heterozygosity and on summary statistics of polymorphism and divergence in D. melanogaster. (A) Genomic location has a significant effect on per-site silent-site heterozygosity (PMWU < 0.001 for all pairwise contrasts), consistent with predicted differences in the effective population size (Ne). The ratio of median mitochondrial to autosomal silent site heterozygosity was 0.157, less than predicted for neutral sites if mitochondrial Ne is one quarter that of the autosomes. mitochondrial DNA (mtDNA), X-chromosome, and autosome data sets contained 12, 1255, and 8073 genes, respectively. (B, C) Distributions of neutrality index (NI) and Z* are similar between mitochondrial and autosomal (abbreviated as A) genes, with moderately significant differences between mitochondrial and X-linked (abbreviated as X) genes. The four mtDNA boxes represent estimates from the corresponding MK tables in Table S2, B−E that used either D. simulans (Table S2, B−C) or D. yakuba (Table S2, D−E) as the outgroup. Dashed lines represent the neutral expectations for these statistics. Three nuclear loci for which NI exceeded 50 were excluded from (B) to improve visualization. Statistical results are presented in Table S2. mtDNA, X-chromosome, and autosome data sets contained 13, 712, and 5401 genes, respectively.