Fig. 1.
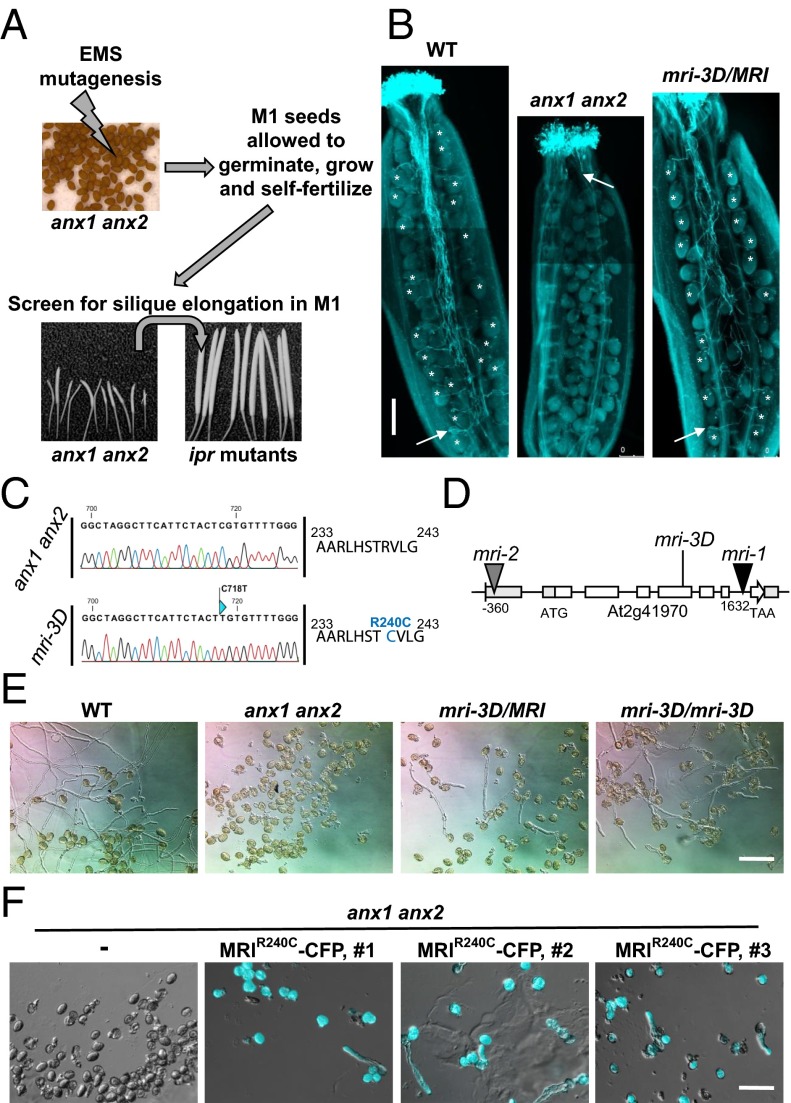
Identification of the ipr mutants, suppressors of anx1 anx2 male sterility, and characterization of maris-3D (mri-3D). (A) Scheme for the anx1 anx2 male sterility-suppressor screen. Reproduced from ref. 6. (B) Aniline blue staining reveals that mri-3D/MRI PTs are able to elongate in vivo and fertilize female gametophytes, unlike anx1 anx2 PTs, which burst and stop growth early in the style. White arrows indicate the tip of the longest PT. Asterisks indicate female gametophytes normally targeted by PTs. (Scale bar: 200 μm.) (C) The mri-3D mutant carries a C718T SNP in AT2G41970 that corresponds to an R240C mutation at the protein level. Shown are spectra from the Sanger sequencing reactions of the AT2G41970 cDNA amplified from anx1 anx2 (Upper) or mri-3D anx1 anx2 (Lower) flowers. (D) AT2G41970 locus with introns, exons, and positions of the mutant alleles. (E) In vitro PT growth assays of WT, anx1 anx2, heterozygous mri-3D/MRI anx1 anx2, and homozygous mri-3D/mri-3D anx1 anx2. Note how the mri-3D mutation partially rescues the anx1 anx2 pollen-bursting phenotype and allows PT growth. See Fig. S2C for corresponding quantification. (Scale bar: 100 μm.) (F) In vitro PT growth assays of untransformed anx1 anx2 and three independent anx1 anx2 transgenic lines expressing MRIR240C-CFP. DIC and CFP channels were overlaid for the transgenic lines. (Scale bar: 80 μm.)