Figure 1. Regulation of the Smoothened GPCR.
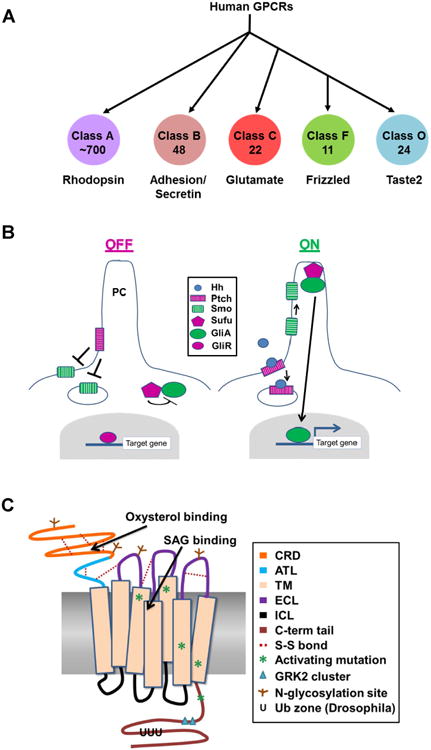
A GPCR classes. Proteins of the GPCR superfamily, which possess 7 TM domains, are grouped into classes based upon their functionality and homology. The diagram indicates human GPCR classes, and denotes the number of proteins found in each class. The largest amount of structural data is available for Class A, the Rhodopsin family. Much of what is known about GPCR topology and activation has been deduced from Class A structural analyses. Smo is a member of the Class F Frizzled family. Classes D (mating pheromone receptors) and E (cAMP receptors) are not found in humans. B. The core Hh pathway. In the absence of Hh ligand, the Hh receptor Ptch localizes to the base of the primary cilium (PC) and inhibits Smo ciliary entry and signaling. Sufu inhibits Gli activator while Gli repressor keeps target genes silenced. Hh binding triggers Ptch internalization, which allows Smo to traffic to the tip of the PC where it signals for Sufu to release Gli activator. Gli activator then translocates to the nucleus to activate target gene expression. Many additional proteins contribute to regulation of these processes. Only the core pathway components are shown here. C. Smoothened functional domains. The cartoon illustrates the distinct Smo functional domains, ligand binding pockets, post-translational modifications and location of known activating mutations. CRD – cysteine rich domain; ATL – amino terminal linker; TM – transmembrane helix; ECL – extracellular loop; ICL – intracellular loop; C-term tail – carboxyl-terminal tail.
