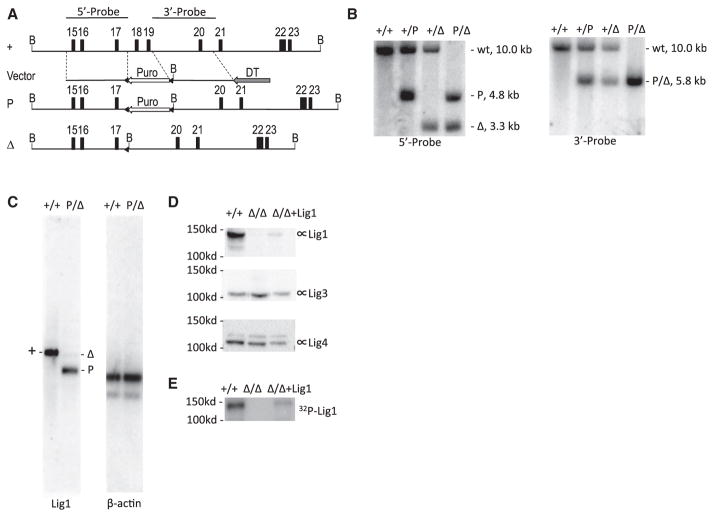
Figure 1. Gene Targeting of Lig1 in CH12F3 Cell Line.
(A) Genomic organization of the wild-type and targeted Lig1 allele. The map is drawn to scale. Exons are indicated by numbered boxes. Arrows indicate transcription orientations of expression cassettes of the puromycin-resistant gene (Puro) and diphtheria toxin A chain (DTA), respectively. B, BamHI restriction sites. Probes used in Southern blot analysis are depicted at the top. A “+” sign indicates the wild-type allele. “P” and “Δ” indicate targeted alleles with or without the Puro cassette, respectively. Triangles indicate loxP sites.
(B) Southern blot analysis of BamHI-digested genomic DNA from wild-type (+/+) and targeted cells (+/P, +/Δ, P/Δ). Genotypes, sizes of bands, and probes are indicated.
(C) Northern blot analysis of polyadenylated RNA. The entire Lig1 coding region sequence was used as a probe. The blot was stripped and stored for a month to allow radioisotype decay before reprobed with β-actin.
(D) Western blot analysis of the Lig1 protein level in wild-type (+/+), Lig1Δ/Δ and mouse Lig1 cDNA complemented Lig1Δ/Δ cells (Δ/Δ+Lig1) with a rabbit polyclonal antibody raised against GST-Lig1 protein (Proteintech 18051-1-AP). The blot was stripped and reprobed with a Lig3 antibody (BD Biosciences, 611876) and Lig4 antibody (Han et al., 2012), respectively, for loading control and protein size distinction.
(E) Adenylation of Lig1 in the cell extract with α-32P-ATP.