Fig. 2.
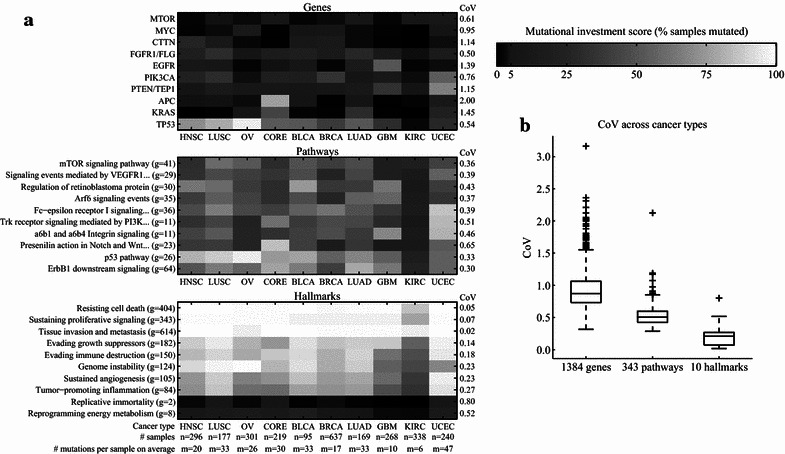
Mutation frequencies across cancer types. a Heatmaps depicting the percentage of samples within a cancer type that have a mutation in a gene (top), a mutation in at least one gene within a pathway (middle), and a mutation in at least one gene that is linked to a hallmark (bottom). Ten genes, 10 pathways, and all 10 hallmarks are shown. These genes and pathways were chosen based on the large variation in their mutation frequencies across cancer types. The coefficient of variation (CoV) in mutation frequencies across cancer types for each depicted gene, pathway, and hallmark is shown to the right of the heatmaps. Some pathway names are shortened for clarity. The number of genes in a pathway is stated in parentheses behind the pathway name. The number of genes linked to a hallmark is stated in parentheses behind the hallmark name. MTOR mechanistic target of rapamycin, MYC v-myc avian myelocytomatosis viral oncogene homolog, CTTN cortactin, FGFR1/FLG fibroblast growth factor receptor 1/filaggrin, EGFR epidermal growth factor receptor, PIK3CA phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha, PTEN/TEP1 phosphatase and tensin homolog/telomerase-associated protein 1, APC adenomatous polyposis coli, KRAS Kirsten rat sarcoma viral oncogene homolog, TP53 tumor protein p53, HNSC head and neck squamous cell carcinoma, LUSC lung squamous cell carcinoma, OV ovarian serous cystadenocarcinoma, CORE colon adenocarcinoma/rectum adenocarcinoma, BLCA bladder urothelial carcinoma, BRCA breast invasive carcinoma, LUAD lung adenocarcinoma, GBM glioblastoma multiforme, KIRC kidney renal clear cell carcinoma, UCEC uterine corpus endometrioid carcinoma. b Boxplot with the CoVs for all genes, pathways, and hallmarks
