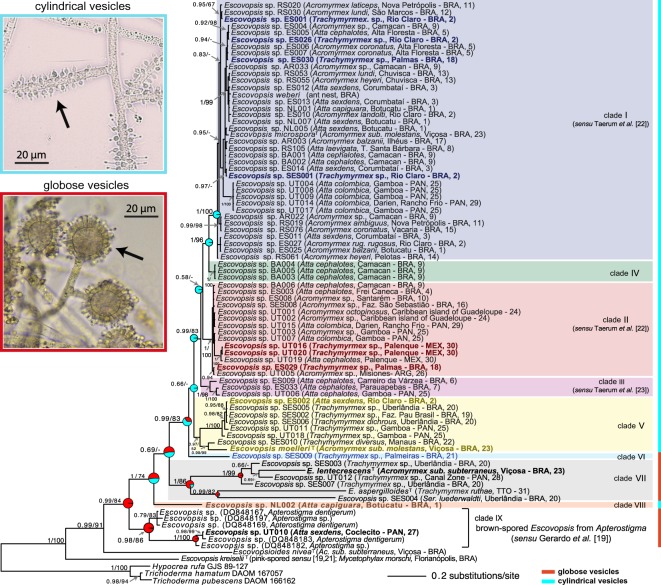
Figure 1.
Phylogeny of Escovopsis associated with higher attine ants based on elongation factor 1-alpha (tef1) and internal transcribed spacer (ITS) markers analysed under Bayesian criteria. Pie-graphs at key nodes show probabilities of ancestral character-states for globose (red) and cylindrical (blue) vesicles. Coloured bars on the right side indicate the observed vesicle morphology, and black arrows point to the respective vesicle structure (400×). The main clades are highlighted in different colours and were consistently supported in both ML and Bayesian analyses. The phylogenetic tree includes also strains from brown-spored Escovopsis associated with Apterostigma ants [19], Escovopsioides nivea and Escovopsis kreiselii (the only species described for the pink-spored Escovopsis, a less derived group that infects lower attine ant gardens, [21]). Three additional species of Hypocreaceae fungi were used as an outgroup. Escovopsis strains in bold indicate exceptions of the co-cladogenesis model (see text). The ant species from which an Escovopsis strain was isolated, its corresponding collection sites and identification (ID) numbers are given in parentheses. GenBank accession number for tef1 and ITS of each strain are available in the electronic supplementary material, table S1. Only posterior probabilities and bootstrap values greater than or equal to 0.5 or 50 are shown. The letter ‘T’ indicates ex-type strains (Ser., Sericomyrmex; sub., subterraneus; ARG, Argentina; BRA, Brazil; MEX, Mexico; PAN, Panama; TTO, Trinidad and Tobago).