Figure 5. Characterization of genomic regions with change of 5hmC in S6KO compared to WT ESCs.
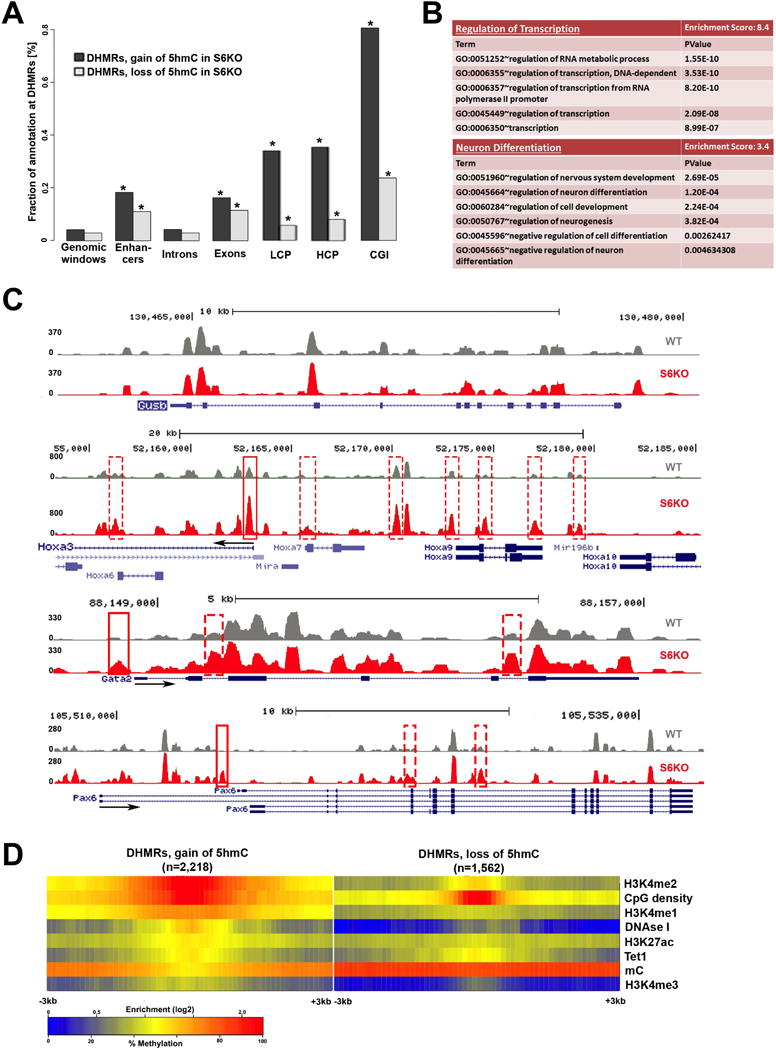
(A) Total DHMRs with gain or loss of 5hmC in S6KO represent 0.04% or 0.03% of the genome, respectively (genomic windows). Both classes of DHMRs are significantly enriched in exons, promoters, and CpG islands, where DHMRs with gain or loss of 5hmC are similarly enriched at exons (p value ≤ 6.3e-155 and p value ≤ 1.5e-110, respectively), whereas DHMRs with gain of 5hmC are much stronger enriched than DHMRs with loss of 5hmC at promoters (p value ≤ 1.3e-252 and p value ≤ 6.3e-16, respectively), and at CpG islands (p value ≤ 1.4e-269 and p value ≤ 1.2e-48, respectively) (Fisher’s exact test).
(B) Functional annotation (Huang et al., 2009) of genes with change of 5hmC (gain or loss) at exons, reveals significantly enriched gene ontology clusters associated with regulation of transcription and neuron differentiation.
(C) UCSC browser visualization of 5hmC of gain of 5hmC in S6KO (red) versus WT (gray) at the promoter of Hoxa3, Gata2 and Pax6 genes, and at multiple other genomic regions in the vicinity. No changes in 5hmC levels on the Gusb gene, is shown as an analytical control.
(D) Enrichment analysis of histone H3 modifications strongly connects H3K4me2 and low methylated regions to DHMRs with gain of 5hmC in S6KO. Both, gain and loss of 5hmC occurs at regions with elevated levels of CpG density and Tet1 binding. The marks are sorted by high (top) to low (bottom) enrichment at the center of DHMRs with gain of 5hmC in S6KO.
