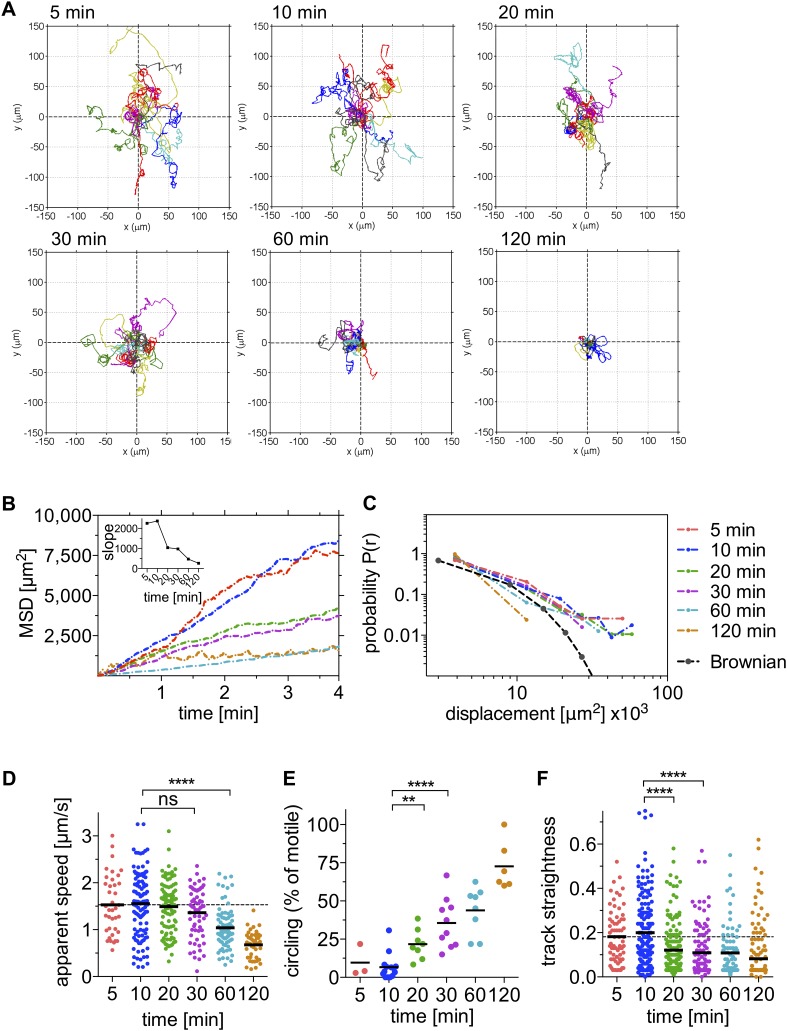
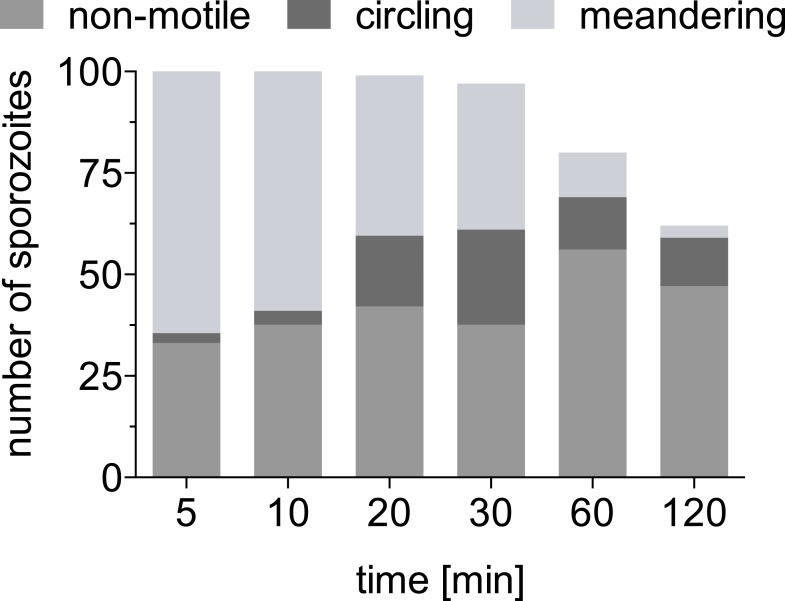
Figure 1. Sporozoite motility is increasingly constrained over time and is not well described by a Brownian walk.
Time-lapse microscopy of sporozoites was started at the indicated time points after intradermal inoculation and 4 min long videos were acquired. See Video 1 for a representative time course. (A) Meandering and linearly moving sporozoites were manually tracked using Imaris software and for each time point 11 representative reconstructed tracks were plotted to a common origin to visualize parasite dispersal over time. For panels B–F, a varying number of videos were processed for each time point after inoculation: 5 min (2 videos/37 tracks), 10 min (14 videos/179 tracks), 20 min (7 videos/95 tracks), 30 min (9 videos/129 tracks), 60 min (8 videos/77 tracks), and 120 min (7 videos/67 tracks). (B) Mean square displacement (MSD) of sporozoite tracks over the duration of the 4 min video, at indicated time points after inoculation, with inset showing the slope obtained through linear regression fitting of MSD curves. (C) The probability distribution P(r) of squared final sporozoite displacements at the end of the 4-min videos. For comparison, the distribution of a Brownian walk is shown. (D) Apparent speed of gliding sporozoites. Bars represent mean values and dashed line marks the mean value at 5 min after inoculation. (E) The percentage of sporozoites gliding continuously in the same circle throughout the duration of the video, showing an increasing proportion of exclusively circling sporozoites at later time points. Every data point represents one video. (F) Straightness of sporozoite tracks, the ratio of displacement to track length of both meandering and linearly moving, as well as continuously circling sporozoites. Bars represent mean values and dashed line marks the mean value at 5 min after inoculation.