Abstract
This study aimed to investigate fungal agents in febrile neutropenic patients with hematological malignancies. Direct microscopy and cultures were performed on clinical samples collected from febrile neutropenic episodes. The galactomannan (GM) antigen was tested using enzyme-linked immunosorbent assays, and Aspergillus fumigatus and Candida albicans deoxyribonucleic acid (DNA) assessed using real-time polymerase chain reaction (PCR) in consecutive serum samples. Of the 199 episodes investigated, 1.5% were classified as definite invasive aspergillosis (IA), 4.0% as IA with high probability, and 4.0% as IA with low probability. Additionally, candidaemia was detected in eight episodes (4.1%). The GM antigen was found negative for 86.4% of episodes, as one positive for 7.0% of episodes, as two or more consecutive positives for 5.5% of episodes, and as positive in any two serum samples in 1.0% of episodes. While no C. albicans DNA was detected in 98.5% of 199 episodes, one positive result was obtained in 1.0% of episodes, and two or more consecutive positives in 0.5% of episodes. A. fumigatus PCR results were found negative in 81.9% of episodes, as one positive in 16.1% of episodes, as positive in any two serum samples in 1.0% of episodes, and consecutively positive in 1.0% of episodes. GM antigen tests were found consecutively positive in all three patients diagnosed as having definite IA. These findings indicate that conventional, serological, and molecular methods should be used in combination to detect fungal agents in febrile neutropenic patients.
KEYWORDS: Febrile neutropenia, invasive fungal infection, culture, PCR, galactomannan
INTRODUCTION
Fungal infections are among the most important causes of mortality and morbidity in neutropenic patients. The risk of development of an invasive fungal infection (IFI) is 15–25% in high-risk patient groups, particularly those with a long history of neutropenia, organ damage, and prior fungal infection or colonization in hematological cancer or transplant cases [1]. Over 90% of fungal infections are caused by Candida and Aspergillus species; however, there has been an increase in fungal infections caused by rarer species, including Trichosporon, Pseudellescheria, Fusarium, and Scedosporium [2].
There is a high mortality rate in neutropenic patients with fungal infections, e.g. 50% with Candida infections [3], and 100% with Aspergillus infections [4]. Previous studies showed that early treatment of fungal infections leads to better prognosis in neutropenic patients. Therefore, early diagnosis and treatment of these infections are of crucial importance. To date, an established diagnostic method for early diagnosis of IFIs, with adequate sensitivity and specificity, is lacking [5]. Diagnosis is based on the fundamental principle of combining clinical and laboratory findings. Methods using direct microscopy and culture are gold standards and remain as important diagnostic tools. However, failure to adequately apply invasive diagnostic methods in certain cases, for example, in the absence of clinical findings and the presence of thrombocytopenia in patients with hematological malignancy, together with long waiting times before culture results are obtained, leads to significant delays in initiating treatment. Therefore, the availability of appropriate tests for early diagnosis is essential in order to guide treatment, particularly in critically ill patients. The serological detection of fungal antigens and the use of molecular methods to test for nucleic acids have led to important advances in diagnosis ability [6]. Among the most studied methods are the galactomannan (GM) antigen test, the 1,3-β-D-glucan (BDG) test, and molecular polymerase chain reaction (PCR) tests [5].
This study aimed (i) to investigate fungal agents in febrile neutropenic patients at high risk of IFIs, by using various methods: direct microscopy, culture, GM antigen test, and real-time PCR, and (ii) to compare the different methods used.
MATERIALS AND METHODS
Patients and samples
This study was approved by the Ethics Committee of the Faculty of Medicine at Gaziantep University under Resolution No. 13.03.2012/129. A total of 101 patients who were admitted with the diagnosis of febrile neutropenia to the Hematology Clinic for Internal Medicine at Şahinbey Research and Application Hospital of Gaziantep University were enrolled in the study. The patients were recruited between April 2012 and July 2013. A majority of 64 patients (63.4%) were female, and 37 (36.6%) were male; the mean age of the study population was 44.6 ± 16.8 years. Criteria for study inclusion were adult patients with a hematological malignancy, a neutrophil count of ≤ 500/mm3, febrile status with temperature of ≥ 38.3°C measured either orally or in the axilla, and the use of chemotherapeutic drugs. All patients received antifungal prophylaxis, according to the recommended guidelines from the European Conference on Infections in Leukemia (ECIL)-3 [7]. A total of 49 patients (48.5%) had acute myelocytic leukemia, 13 (12.9%) acute lymphocytic leukemia, 10 (9.9%) myeloma, eight (7.9%) diffuse large B-cell lymphoma, three (3.0%) myelodysplastic syndrome, three (3.0%) a lymphoma of the central nervous system, three (3.0%) aplastic anemia, three (3.0%) mantle cell lymphoma, two (2.0%) follicular lymphoma, two (2.0%) peripheral T-cell lymphoma, one (1.0%) anaplastic large cell lymphoma, one (1.0%) Hodgkin’s lymphoma, one (1.0%) chronic lymphocytic leukemia, one (1.0%) angioimmunoblastic lymphoma, and one (1.0%) Burkitt’s lymphoma. In addition, only 42 out of the 101 patients (41.6%) underwent peripheral blood stem cell transplantation (PBSCT). Of those 42 patients, 20 patients (47.6%) were administered allogeneic PBSCT from human leucocyte antigen (HLA)-identical siblings (47.6%), 21 patients (50%) autologous PBSCT, and one patient (2.4%) autologous plus allogeneic PBSCT. For each patient, a minimum of one and a maximum of five neutropenic episodes were recorded and investigated for IFIs, as follows: one episode in 101 patients, two episodes in 61 patients, three episodes in 29 patients, four episodes in seven patients, and five episodes in one patient.
All clinical samples were collected for subsequent laboratory analyses, including the GM antigen test, and Aspergillus fumigatus and Candida albicans PCR tests, from 194 neutropenic episodes for blood culture, 61 episodes for urine culture, 34 episodes for sputum culture, 12 episodes for catheter culture, two episodes for tracheal aspirate (TRA), and one episode for bronchoalveolar lavage (BAL), throat swab, wound swab, brain biopsy material, and pleural material cultures. The number of clinical samples sent for culture were as follows: 443 blood samples, 229 urine samples, 40 sputum samples, 20 catheter samples, two TRA samples, a BAL sample, a throat swab sample, a wound swab sample, a brain biopsy material, and a pleural sample.
A total of 813 serum samples from 199 episodes in 101 febrile neutropenic patients were screened for the GM antigen, with an average of five serum samples analyzed on two consecutive occasions within a week from each episode, using the enzyme-linked immunosorbent assay (ELISA) method. A. fumigatus DNA was investigated in 818 plasma samples and C. albicans DNA in 749 plasma samples from 199 episodes with the method of real-time PCR.
The study patients were grouped, according to the European Organization for Research and Treatment of Cancer (EORTC) criteria [8].
Direct microscopic examination and culture
Samples collected from the respiratory tract, wound swabs, and brain biopsy were analyzed using direct microscopy. All samples were treated with 10% potassium hydroxide solution and examined for fungal infection under a light microscope (Olympus, Tokyo, Japan), using objectives ×20 to ×40.
All clinical samples, except blood culture samples, were cultured on 5% sheep blood agar (bioMérieux, Marcy-l’Etoile, France) and eosin–methylene blue (EMB) agar (bioMérieux, Marcy-l’Etoile, France) media, as well as two Sabouraud dextrose agar (SDA) (bioMérieux, Marcy-l’Etoile, France) media. A colony count of 104 cfu/mL in urine cultures was considered as significant growth [9], and colonies obtained from all cultures were Gram-stained. Gram-positive cream-colored and yellow colonies, which also produced a distinctive yeast smell, were further examined microscopically with the germ tube (germination pipe) test on Cornmeal-Tween 80 agar medium (Oxoid, Hampshire, England), and were also identified, according to their biochemical properties, using the Vitek 2 automated system (bioMérieux, Durham, St. Louis, USA). C. albicans ATCC 14053 and C. lusitaniae ATCC 34449 strains were used as positive identification controls.
Blood culture
Blood cultures demonstrating growth in the BacTAlert 3D (bioMérieux, Durham, St. Louis, USA) automated blood culture system on 5% sheep blood agar (bioMérieux, Marcy-l’Etoile, France) and EMB agar (bioMérieux, Marcy-l’Etoile, France) media were incubated for 24–48 hours. A passage was performed from cultures producing Gram-positive yeast cells detected on the SDA medium. Cream-colored and yellow colonies with a distinctive yeast smell at the end of incubation period were identified, using the conventional as well as the Vitek 2 (bioMérieux, Durham, St.Louis, USA) automated systems.
Serological tests
Serum samples were assessed for the presence of the GM antigen by ELISA using the Platelia Aspergillus kit (BioRad, Marnes-la-Coquette, France), according to the manufacturer’s instructions.
A GM index of < 0.5 was considered to be negative, whereas a GM index of ≥ 0.5 was considered positive.
Molecular methods
The MagNA Pure LC kit (Roche Diagnostics, Mannheim, Germany) was used for nucleic acid isolation in all plasma samples collected, according to the manufacturer’s instructions. The isolated deoxyribonucleic acid (DNA) samples were then analyzed by real-time PCR, in order to detect A. fumigatus and C. albicans DNA, using the LightMix kit (Tib Molbiol, Berlin, Germany) in a Light Cycler 2.0 apparatus (Roche Diagnostics, Mannheim, Germany), according to the manufacturer’s instructions.
The LightMix kit detects the fungal 18S ribonucleic acid (RNA) region that shows the presence of fungal DNA in a nucleic acid extraction. A LightMix Kit C. albicans (Tib Molbiol, Berlin, Germany) has a 490 bp long fragment of the C. albicans genome was amplified with specific primers and detected with probes labelled with LightCycler Red 640 (Tib Molbiol, Berlin, Germany). LightMix Kit A. fumigatus (Tib Molbiol, Berlin, Germany) has a 504 bp long fragment of the Aspergillus fumigatus genome and was amplified with primers, and detected and labelled similarly. The kits also include control DNAs of C. albicans and A. fumigatus equivalent to 105 targets. Approximately 10 C. albicans and A. fumigatus DNA copies were assessed, using LightCycler 1.×/2.0 Apparatus and LightCycler FastStart DNA Master HybProbe (Roche Diagnostics, Mannheim, Germany). The positive control used was provided in the manufacturer’s kit, whereas water was used as the negative control. The resulting fluorescent curves were evaluated with LightCycler software version 4.1 (Roche Diagnostics, Mannheim, Germany).
Histopathological assessments
Results of histopathological assessments of brain biopsy samples performed on three patients were retrieved from the patient files.
Statistical analyses
Correlation among the categorical variables was tested using the chi-square analysis. Specificity values and 95% confidence intervals (CIs) were calculated for the various methods used and compared with the gold standard, which is to culture for Candidas and also galactomannan antigen for A. fumigatus according to EORTC criteria [8]. Furthermore, the agreement between methods was checked using the kappa statistic. Frequency, percentage, and mean ± standard deviation (SD) values were provided as descriptive statistics. The SPSS for Windows version 11.5 package program (SPSS Inc., Chicago, Illinois, USA) was used for statistical analyses, and P < 0.05 was considered statistically significant. All statistical evaluations made use of the number of neutropenic episodes in patients, and not the number of patients or samples.
RESULTS
Of a total of 199 neutropenic episodes investigated, according to the classification by the EORTC/Mycoses Study Group, definite invasive aspergillosis (IA) was detected in three (1.5%) episodes by histopathological observation of hyphal structures with septae in brain biopsy samples (data not shown); eight (4.0%) neutropenic episodes were identified as high-probability IA, and eight (4.0%) as low-probability IA, whereas the remaining 180 (90.5%) episodes were not found at risk of IA. Moreover, candidaemia was detected in eight episodes (4.1%).
Direct microscopic examination and culture
Direct microscopy of clinical samples revealed no fungal structures. The distribution of culture results from the clinical samples, according to the number of neutropenic episodes, is shown in Table 1.
TABLE 1.
Distribution of culture results for patient samples analysed, according to the number of episodes
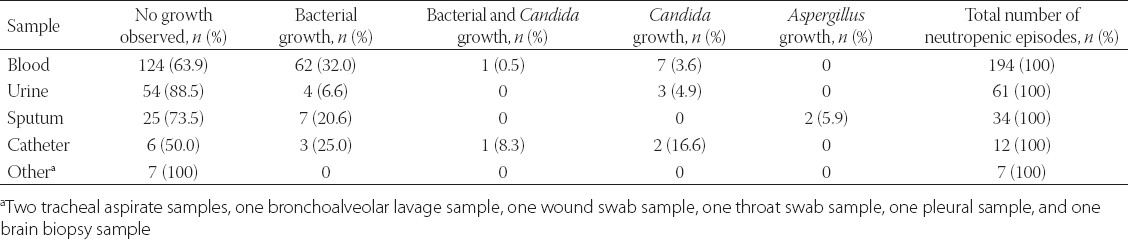
A total of 443 blood cultures, collected from 194 febrile neutropenic episodes in 101 patients, were analyzed. No growth occurred in blood cultures from 124 (63.9%), out of 194, episodes, whereas bacterial growth was detected from 62 (32%) episodes and the growth of various Candida species from seven (3.6%) episodes. Of the Candida species that grew, four were identified as C. glabrata, and one was identified as C. parapsilosis, one as C. albicans, and one as C. tropicalis. On the other hand, mixed infection with both C. parapsilosis and a bacterial agent was detected from one (0.5%) neutropenic episode.
Furthermore, a total of 229 urine samples collected from 61 episodes were analysed. Of the Candida species that grew, two were identified as C. glabrata, and one as C. albicans.
Additionally, a total of 40 sputum cultures from 34 neutropenic episodes were assessed. No growth was detected from 25 (73.5%), out of 34, episodes, whereas bacterial growth was obtained from seven (20.6%) episodes. In addition, growth of Aspergillus species was observed from two episodes (5.9%), although growth was found in only a single culture, with no further Aspergillus growth obtained in subsequent sputum cultures from the patients.
Moreover, a total of 20 catheter cultures were analyzed from 12 episodes. No growth occurred in six (50%), of the 12, episodes, while bacterial growth was found from three episodes (25%) and Candida species from two episodes (16.6%), although both Candida species and a bacterial agent grew concurrently from one of these two episodes (8.3%). The Candida species were identified as C. albicans from two episodes, and as C. tropicalis from one episode.
GM antigen findings
The GM antigen was found negative for 86.4% of the episodes, as one positive for 7.0% of them, two or more consecutive positives for 5.5% of them, and positive in any two sera in 1.0% of them. The distribution of the GM results obtained with a GM index of ≥ 0.5 is shown in Table 2.
TABLE 2.
Distribution of galactomannan results obtained with index values of ≥0.5

PCR findings
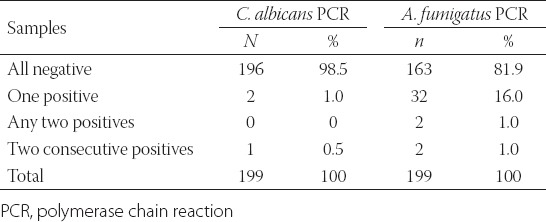
A total of 749 plasma samples to assess for C. albicans DNA, and 818 plasma samples to check for A. fumigatus DNA, were analysed from 199 neutropenic episodes in 101 febrile neutropenic patients, twice a week from each episode. While no C. albicans DNA was determined in 98.5% of 199 episodes, one positive was determined in 1.0% of them and two or more consecutively positives were determined in 0.5% of them. A. fumigatus PCR results were found negative in 81.9% of the episodes, as one positive in 16.1% of them, positive in any two sera in 1.0% of them, and consecutively positive in 1.0% of them. C. albicans and A. fumigatus PCR results from all episodes are presented in Table 3.
TABLE 3.
C. albicans and A. fumigatus PCR results from all neutropzc episodes
Comparison of culture and PCR results
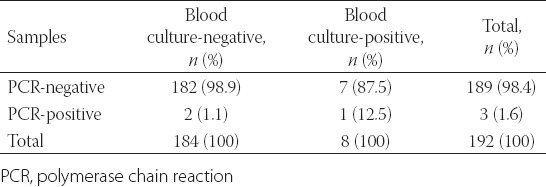
Blood cultures from 194 episodes were analysed, and C. albicans PCRs were carried out simultaneously from 192 of the 194 episodes. A comparison between the blood culture results and C. albicans PCR results is shown in Table 4. In addition, the specificity and negative predictive value (NPV) of the PCR were 98.9% (95% CI 96.2–99.9) and 96.8% (95% CI 93.2–98.8), respectively. Of the Candida species that grew in blood cultures, four were identified as C. glabrata and two as C. parapsilosis, whereas one was identified as C. albicans and one as C. tropicalis. It was noted that the C. albicans strain that grew in the blood culture was also detected with C. albicans PCR.
TABLE 4.
Comparison between blood culture results and C. albicans PCR results
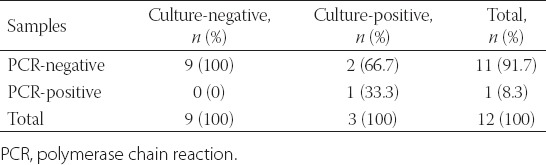
Moreover, a total of 20 catheter cultures from 12 episodes were assessed. Candida species grew from three episodes (25%), with C. albicans identified from two episodes and C. tropicalis from one episode. On comparing C. albicans PCR results with the catheter culture results, the specificity and NPV of the PCR were 100% (95% CI 66.4–100) and 81.8% (95% CI 48.2–97.7), respectively. The comparison between the catheter culture results and C. albicans PCR results is presented in Table 5.
TABLE 5.
Comparison between catheter culture results and C. albicans PCR results
The growth of Candida species was observed from three of 61 episodes for which urine cultures were analysed. C. glabrata was identified from two episodes, and C. albicans identified from one episode. It was noted that C. albicans had also grown in the blood and catheter cultures of the patient in whose urine culture C. albicans had grown. When C. albicans PCR results were compared with the urine culture results, the specificity and NPV of PCR were 98.3% (95% CI 90.8–99.9) and 96.6% (95% CI 88.3–99.6), respectively. It was observed that the C. albicans strain that had grown in the urine culture was also detected on C. albicans PCR.
Comparison of A. fumigatus PCR and GM results
A comparison of the results obtained with a GM index of ≥ 0.5 and A. fumigatus PCR results is provided in Table 6.
TABLE 6.
Comparison between results achieved with galactomannan index values ≥0.5 and A. fumigatus PCR results
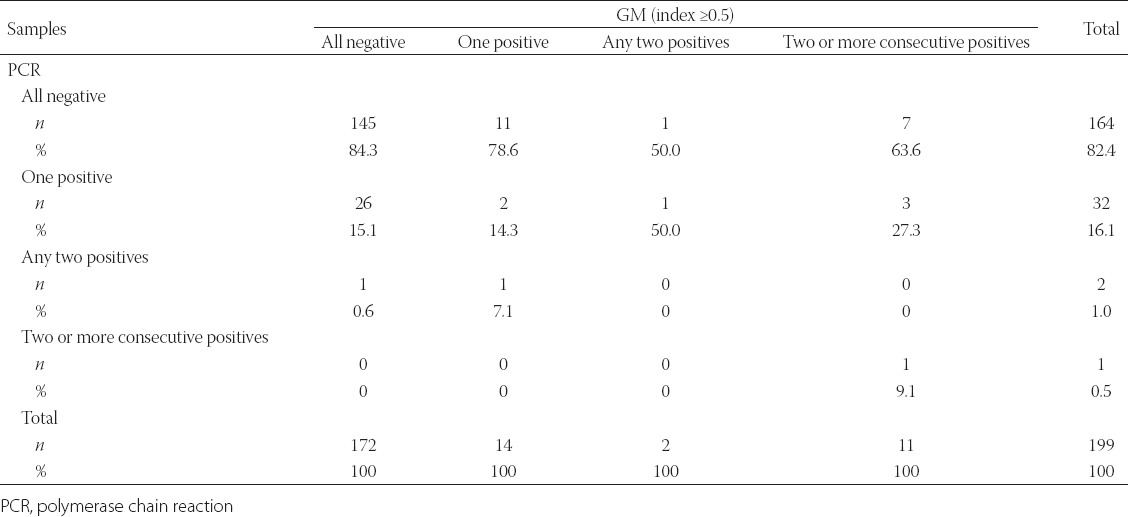
On comparing A. fumigatus PCR and GM results, with a single positive value considered as significant, the specificity and NPV of PCR were 84.3% (95% CI 78.0–89.4) and 88.4% (95% CI 82.5–92.9), respectively. When the two methods were evaluated for agreement, poor agreement was observed between the tests (kappa = 0.124, p = 0.077).
In addition, on comparing any positive results from A. fumigatus PCRs with consecutive positive GM results, the specificity and NPV of the PCR were 83.87% (95% CI 77.78–88.85) and 95.12% (95% CI 90.61–97.87), respectively, and poor agreement was found between the two tests (kappa = 0.125, p = 0.041).
Comparison of histopathological results with GM and A. fumigatus PCR results
No fungal growth was detected in the culture of a brain biopsy sample of one of three patients, although hyphal structures with septae were detected histopathologically in the brain biopsy sample; no culture had been carried out on brain biopsy samples from the other two patients. Consecutive positive GM results were obtained in all three patients (100%). A. fumigatus PCR results were negative in two (66.7%) of these patients, whereas consecutive positive PCR results were obtained in one patient (33.3%). The sensitivity and specificity of the tests could not be calculated and compared, as histopathological examinations, although the gold standard, could not be performed for all study patients.
DISCUSSION
Superficial and deep opportunistic fungal infections are frequent in neutropenic oncology patients and can be caused by any fungus, with the commonest fungi being from the Candida genus, followed by Aspergillus [10-12]. While the commonest agent is C. albicans in candidaemia, the incidence of non-albicans species is also increasing [13-16].
In a meta-analysis performed for Invasive Candidiasis (IC) [17], PCR sensitivity and specificity were reported as 100% in patients with candidaemia, whereas PCR sensitivity and specificity were 95% and 92%, respectively, in suspected cases of IC. In contrast, in cases of proven and highly probable IC, blood cultures and PCR had a positivity rate of 38% and 85%, respectively. In a study by Nguyen et al. [18] investigating the diagnostic values of real-time PCR, BDG, and blood cultures in IC cases, PCR was reported to be more sensitive than the other diagnostic methods. In this study, it was stated that identical Candida species were detected in 82% of patients by PCR and blood cultures and sensitivity was 98% when blood cultures and PCR were used in combination. In a study carried out using real-time PCR [19], it was reported that DNA amplification was 100% specific for Candida species. In our study, analysis for C. albicans DNA was found as one positive in 1% of episodes, and consecutively positive in 0.5% of episodes. The real-time PCR kit which we used for nucleic acid amplification can only detect C. albicans DNA. It was noted that the C. albicans strain that grew in the blood culture was also detected with C. albicans PCR. Also non-albicans Candida species can often be the infecting agents in patients with candidaemia. Therefore, it is important to select a PCR kit for detecting also non-albicans Candida spp.
In general, candiduria very rarely causes candidaemia in immune-compromised patients [20]. In our study, the growth of C. albicans was detected in blood and catheter cultures of a patient whose urine culture also showed growth of C. albicans. As a result C. albicans DNA was found on this patient. Growth of C. albicans in the urine culture of one of our patients and the positive C. albicans-specific PCR result of the same patient suggests that Candida growth in urine cultures might be significant in neutropenic patients, and that it merits careful evaluation.
IA is a serious disease, which in the majority of cases can be fatal unless diagnosed and treated early [21]. Although culture is the gold standard for a definitive diagnosis, it is difficult and time-consuming to carry out [22]. Also in our study, the number of samples collected, such as BALs or TRAs, was quite limited, due to the difficulty in obtaining invasive respiratory tract samples from our study patients. GM antigen, BDG, and PCR tests are the most widely studied methods [23]. Studies using the platelia GM enzyme immunoassay (EIA) test yielded positive results, and the sensitivity and specificity varied between 29% and 99% in many prospective studies [24]. In a meta-analysis of 27 studies by Pteiffer et al. [25], the sensitivity and specificity of the GM test were found to be 71% and 89%, respectively. In our study, the GM antigen test was obtained as one positive during 7.03% of episodes, as two or more consecutive positives during 5.52% of episodes, and was positive in any two serum samples in 1% of episodes GM antigen tests were found to be consecutively positive in all three patients with a confirmed diagnosis of IA. Although it is difficult to differentiate fungal colonization from an infection in immunosuppressed patients, it is recommended to screen for antigens twice weekly in serum samples of at-risk patients, as colonization can rapidly turn into an infection, with a tendency to become widespread in the body [26]. However, in their recent study, Duarte et al. [27] reported that, in patients receiving posaconazole prophylaxis, the GM results might be misleading, with markedly high false-positive rates and decreased PPV rates, thus failing to show true positive results. The low pretest risk of IA, in the context of effective antifungal prophylaxis, renders serum GM surveillance of asymptomatic patients unreliable, as all results would be either negative or false positive. However, the test remains useful in the diagnosis of patients with a clinical suspicion of invasive fungal disease, thus calling for a more efficient co-utilisation of effective antifungal prophylaxis and GM testing in this clinical setting.
Indeed, molecular methods are important alternatives to conventional methods, as they are sensitive and safe and can yield fast results [28]. In a study by Silva et al. [29], IA was investigated, using PCR, in 1311 consecutive serum samples from 172 neutropenic patients, and by screening for the GM antigen in 806 consecutive serum samples from 169 neutropenic patients. The authors reported the sensitivity and specificity to be 75.0% and 91.9%, respectively, for PCR and 87.5% and 93.1%, respectively, for GM. Thus, it was suggested that both tests were important in the diagnosis of IA in two or more consecutive serum samples. Other similar studies also recommended the combined use of GM and PCR tests, with emphasis on increased sensitivity [30-32]. In our study, the specificity and NPV were 84.3% and 88.4%, respectively, when we compared A. fumigatus PCR and GM results, with a single positive value considered significant. Furthermore, when we compared any positivity in A. fumigatus PCR with the consecutive positives in GM, the specificity and NPV of the PCR were 83.9% and 95.1%, respectively. GM is an immunogenic antigen, and its presence in A. fumigatus and other Aspergillus species (A. flavus, A. niger, A. versicolor, A. terreus, A. nidulans, and A. oryzae) has been shown [33]. In our study, we believe that the real-time PCR kit used influenced the sensitivity of the test, as it was specific only to A. fumigatus.
CONCLUSION
The use of conventional diagnostic methods, such as direct microscopy and culture, as well as serological and molecular methods alone is inadequate in the diagnosis of fungal infections in patients with immunodeficiency. These methods should be used in combination. The limitations of our study is including routine patient’s samples which have been set to routine mediums. Our study just has created for evaluating of using methods that determines fungal agents. Therefore we’ve not added bacterial agents to discussion.
ACKNOWLEDGEMENTS
We thank the patients of the Şahinbey Research and Application Hospital of Gaziantep University for their participation in this study. This study was presented with an oral presentation award at the first National Congress of Medical Mycology (24–26 September 2014, Ankara, Turkey).
DECLARATION OF INTERESTS
The authors declare no conflict of interest.
REFERENCES
- [1].O’Brien SN, Blijlevens NM, Mahfouz TH, Anaissie EJ. Infections in patients with hematological cancer: recent developments. Hematology Am Soc Hematol Educ Program. 2003:438–472. doi: 10.1182/asheducation-2003.1.438. doi: http://dx.doi.org/10.1182/asheducation-2003.1.438 . [DOI] [PubMed] [Google Scholar]
- [2].Richardson M, Lass-Flörl C. Changing epidemiology of systemic fungal infections. Clin Microbiol Infect. 2008;14(4):5–24. doi: 10.1111/j.1469-0691.2008.01978.x. doi: http://dx.doi.org/10.1111/j.1469-0691.2008.01978.x . [DOI] [PubMed] [Google Scholar]
- [3].Viscoli C, Girmenia C, Marinus A, Collette L, Martino P, Vandercam B, et al. Candidemia in cancer patients: A prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC) Clin Infect Dis. 1999;28(5):1071–079. doi: 10.1086/514731. DOI: http://dx.doi.org/10.1086/514731 . [DOI] [PubMed] [Google Scholar]
- [4].Abbasi S, Shenep JL, Hughes WT, Flynn PM. Aspergillosis in children with cancer: A 34-year experience. Clin Infect Dis. 1999;29(5):1210–1219. doi: 10.1086/313445. doi: http://dx.doi.org/10.1086/313445 . [DOI] [PubMed] [Google Scholar]
- [5].Odabaşı Z. Early diagnosis in invasive fungal infections. The 3rd Postgraduate Training Course on Febrile Neutropenia. Ankara. 2004:48–53. [Google Scholar]
- [6].Arda B. Definitions related to radiologic and serologic tests for diagnosis of invasive fungal infections. ANKEM Derg. 2009;23:122–125. [Google Scholar]
- [7].Maertens J, Marchetti O, Herbrecht R, Cornely OA, Flückiger U, Frere P, et al. European guidelines for antifungal management in leukemia and hematopoietic stem cell transplant recipients: summary of the ECIL 3—2009 Update. Bone Marrow Transplant. 2011;46(5):709–718. doi: 10.1038/bmt.2010.175. DOI: http://dx.doi.org/10.1038/bmt.2010.175 . [DOI] [PubMed] [Google Scholar]
- [8].De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, et al. Revised Definitions of Invasive Fungal Disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46(12):1813–1821. doi: 10.1086/588660. doi: http://dx.doi.org/10.1086/588660 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kauffman CA, Fisher JF, Sobel JD, Newman CA. Candida urinary tract infections diagnosis. Clin Infect Dis. 2011;52(6):452–456. doi: 10.1093/cid/cir111. doi: http://dx.doi.org/10.1093/cid/cir111 . [DOI] [PubMed] [Google Scholar]
- [10].De Pauw BE, Verweıj PE. Infections in patients with acute hematologic malignancies. In: Mandell GL, Bennet JE, Dolin R, editors. Principles and Practice of Infectious Diseases. Philadelphia: Churchill Livingstone; 2005. pp. 3432–3441. [Google Scholar]
- [11].A working group on febrile neutropenia. Guidelines for Diagnosis and Therapy in Febrile Neutropenic Patients. FLORA. 2004;9(1):5–28. [Google Scholar]
- [12].Metan G. A road map in febrile neutropenia: diagnostic approaches. EKMUD scientific platform. Antalya. 2013:87–88. [Google Scholar]
- [13].Atalay MA, Sav H, Demir G, Koç NA. Distribution of Candida Species Isolated From Blood Cultures and in- Vitro Susceptibilities to Amphotericin B and Fluconazole. Selçuk Tıp Dergisi. 2012;28(3):149–151. [Google Scholar]
- [14].Şahin E, Ersöz G, Otağ F, Kandemir Ö, Tiftik N, Kaya A, et al. Evaluation of Candida spp. Isolated from febrile neutropenic patients with hematological malignancies. Turkish Journal of Infection. 2006;20(2):121–124. [Google Scholar]
- [15].Kaya D, Kaptanoğlu S, Üstüner Z, Ertör E. Typing of yeasts isolated from the specimens of neutropenic patients and investigation of fluconazole resistance. KLİMİK Derg. 2001;14(1):14–16. [Google Scholar]
- [16].Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. doi: 10.1086/421946. doi: http://dx.doi.org/10.1086/421946 . [DOI] [PubMed] [Google Scholar]
- [17].Avni T, Leibovici L, Paul M. PCR diagnosis of invasive Candidiasis: Systematic review and meta-analysis. J Clin Microbiol. 2011;49(2):665–670. doi: 10.1128/JCM.01602-10. DOI: http://dx.doi.org/10.1128/JCM.01602-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nguyen MH, Wissel MC, Shields RK, Salomoni MA, Hao B, Press EG, et al. Performance of Candida real-time Polymerase Chain Reaction, B-D-Glucan assay, and Blood cultures in the diagnosis of invasive candidiasis. Clin Infect Dis. 2012;54(9):1240–1248. doi: 10.1093/cid/cis200. doi: http://dx.doi.org/10.1093/cid/cis200 . [DOI] [PubMed] [Google Scholar]
- [19].Klingspor L, Jalal S. Molecular detection and identification of Candida and Aspergillosis spp. from Clinical Samples Using Real Time PCR, Clin Microbiol Infect. 2006;12(8):745–753. doi: 10.1111/j.1469-0691.2006.01498.x. doi: http://dx.doi.org/10.1111/j.1469-0691.2006.01498.x . [DOI] [PubMed] [Google Scholar]
- [20].Uzun Ö. An approach to fungal infections in the intensive care unit. The Intensive Care Journal. 2003;3(2):135–144. [Google Scholar]
- [21].Maertens J, Van Eldere J, Verhaegen J, Verbeken E, Verschakelen J, Boogaerts M. Use of circulating galactomannan screening for early diagnosis of invasive aspergillosis in allogeneic stem cell transplant recipients. J Infect Dis. 2002;186(9):1297–1306. doi: 10.1086/343804. doi: http://dx.doi.org/10.1086/343804 . [DOI] [PubMed] [Google Scholar]
- [22].Perfect JR, Cox GM, Lee JY, Kauffman CA, de Repentigny L, Chapman SW, et al. The Mycoses Study Group. The impact of culture isolation of Aspergillus species: a hospital-based survey of aspergillosis. Clin Infect Dis. 2001;33(11):1824–1833. doi: 10.1086/323900. doi: http://dx.doi.org/10.1086/323900 . [DOI] [PubMed] [Google Scholar]
- [23].Odabaşı Z. Imaging methods and serodiagnosis in invasive fungal infections. In: Arman D, Odabaşı Z, editors. Fungal Infections and their Treatment, a series of treatments in infectious diseases-12 2009. Ankara: Scientific Medicine Press; 2009. pp. 19–27. [Google Scholar]
- [24].Demirhan Delibalta G, Gençer S, Çağ Y, Özer S. The Value of Galactomannan Test in the Evaluation of Invasive Aspergillosis in Patients with Prolonged Febrile Neutropenia. FLORA. 2012;17(1):11–17. [Google Scholar]
- [25].Pfeiffer CD, Fine JP, Safdar N. Diagnosis of invasive aspergillosis using a galactomannan assay: a meta-analysis. Clin Infect Dis. 2006;42(10):1417–1427. doi: 10.1086/503427. doi: http://dx.doi.org/10.1086/503427 . [DOI] [PubMed] [Google Scholar]
- [26].Kantarcıoğlu AS, Yücel A. Aspergillus species and invasive aspergillosis: Mycology, pathogenesis, laboratory diagnosis, resistance of antifungal agents and susceptibility tests. Cerrahpaşa J Med. 2003;34(3):140–157. [Google Scholar]
- [27].Duarte RF, Sánchez-Ortega I, Cuesta I, Arnan M, Patino B, Fernandez de Sevilla A, et al. Serum galactomannan-based early detection of invasive aspergillosis in hematology patients receiving effective anti-mold prophylaxis. Clin Infect Dis. 2014;59(12):1696–1702. doi: 10.1093/cid/ciu673. doi: http://dx.doi.org/10.1093/cid/ciu673 . [DOI] [PubMed] [Google Scholar]
- [28].Wengenack NL, Binnicker MJ. Fungal molecular diagnostics. Clin Chest Med. 2009;30(2):391–408. doi: 10.1016/j.ccm.2009.02.014. doi: http://dx.doi.org/10.1016/j.ccm.2009.02.014 . [DOI] [PubMed] [Google Scholar]
- [29].Lopes da Silva R, Ribeiro P, Abreu N, Ferreira T, Fernandes T, Monteiro A, et al. Early diagnosis of invasive Aspergillosis in neutropenic patients. Comparison between serum Galactomannan and Polymerase Chain Reaction. Clinical Medicine Insights Oncol. 2010;4:81–88. doi: 10.4137/cmo.s5228. DOI: http://dx.doi.org/10.4137/CMO.S5228 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Challier S, Boyer S, Abachin E, Berche P. Development of a Serum-Based Taqman Real-Time PCR Assay for Diagnosis of Invasive Aspergillosis. J Clin Microbiol. 2004;42(2):844–846. doi: 10.1128/JCM.42.2.844-846.2004. DOI: http://dx.doi.org/10.1128/JCM.42.2.844-846.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sanguinetti M, Posteraro B, Pagano L, Pagliari G, Fianchi L, Mele L, et al. Comparison of Real-Time PCR, Conventional PCR, and Galactomannan Antigen Detection by Enzyme-Linked Immunosorbent Assay Using Bronchoalveolar Lavage Fluid Samples from Hematology Patients for Diagnosis of Invasive Pulmonary Aspergillosis. J Clin Microbiol. 2003;41(8):3922–3925. doi: 10.1128/JCM.41.8.3922-3925.2003. DOI: http://dx.doi.org/10.1128/JCM.41.8.3922-3925.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Aguado JM, Vazquez L, Fernandez-Ruiz M, Villaescusa T, Ruiz-Camps I, Barba P, et al. Serum galactomannan versus a combination of galactomannan and polymerase chain reaction-based Aspergillus DNA detection for early therapy of invasive aspergillosis in high-risk hematological patients: a randomized controlled trial. Clin Infect Dis. 2015;60(3):405–14. doi: 10.1093/cid/ciu833. doi: http://dx.doi.org/10.1093/cid/ciu833 . [DOI] [PubMed] [Google Scholar]
- [33].Wheat LJ. Non culture diagnostic methods for invasive fungal infections. Curr Infect Dis Rep. 2007;9(6):465–471. doi: 10.1007/s11908-007-0071-7. DOI: http://dx.doi.org/10.1007/s11908-007-0071-7 . [DOI] [PubMed] [Google Scholar]


