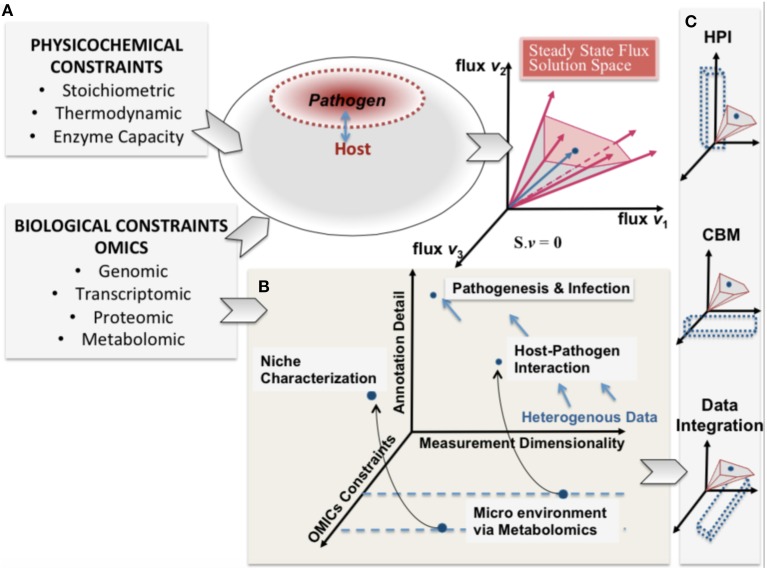
Figure 1.
A conceptual representation of integrating constraint-based modeling and omic data. The heterogeneity of omic data (biological constraints) and their integration is represented in parallel with the phenotypic solution space of the high dimensional host-pathogen model derived from physicochemical constraints. The degree of constraints represented will depend on the measurement capability and also define a reference set of behaviors that are feasible. (A) enumerates the heterogeneity of constraints for both host and pathogen and the resultant mathematically feasible and the potential biologically relevant solution space. In (B) pathogenesis and infection are shown from the perspective of 3 dimensions (i) omics constraints (also determined by experimental constraints) (ii) Annotation detail (based on existing legacy data) and (iii) the measurement dimensionality (also defining dimensionality of data). (C) shows that understanding host-pathogen interaction would be possible at multiple scales by integrating heterogeneous data/measurements and constraint-based modeling algorithms. The opportunity afforded by the legacies of high throughput omics experimentation and systems-level mathematical models would help understand the emergent host-pathogen interaction.