Table 1.
| Space group | P21 | P1 |
|---|---|---|
| Unit-cell parameters (Å°) | a=54.33, b=37.40, c=65.48, α=90, β=98.57, γ=90 | a=35.20, b=62.42, c=66.05, α=72.39, β=74.57, γ=73.63 |
| Resolution (Å) | 15.00–1.98 (2.01–1.98) | 48.68–2.63 (2.70–2.63) |
| Completeness (%) | 99.5 (99.9) | 97.0 (94.3) |
| Redundancy | 3.4 (3.4) | 4.7 (4.4) |
| <I/σ(I)> | 19.1 (2.0) | 14.2 (3.4) |
| Rmerge† | 0.049 (0.58) | 0.088 (0.373) |
| Reflections in working set | 17316 (1142) | 13737 (676) |
| Reflections in test set | 887 (58) | 732 (56) |
| Rwork/Rfree | 0.218/0.247 | 0.235/0.275 |
| Contents of asymmetric unit | 1 Protein:DNA complex | 2 Protein:DNA complexes |
| Number of atoms | ||
| Protein | 756 | 1512 |
| DNA | 814 | 1628 |
| Ligand/ion | 7 | 14 |
| Water | 93 | 0 |
| B factors (Å2) | ||
| Protein | 53 | 62 |
| DNA | 60 | 69 |
| Ligand/ion | 62 | 67 |
| Water | 34 | N/A |
| R.m.s.d bond lengths (Å) | 0.005 | 0.004 |
| R.m.s.d bond angles (°) | 1.08 | 0.92 |
| Ramachandran plot | ||
| Favored (%) | 96.7 | 97.3 |
| Disallowed (%) | 0.0 | 0.0 |
| Refinement program | REFMAC5 | REFMAC5 |
| PDB code | 3VD6 | 3VEK |
Values in parentheses are for highest-resolution shell.
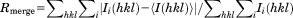 .
.
