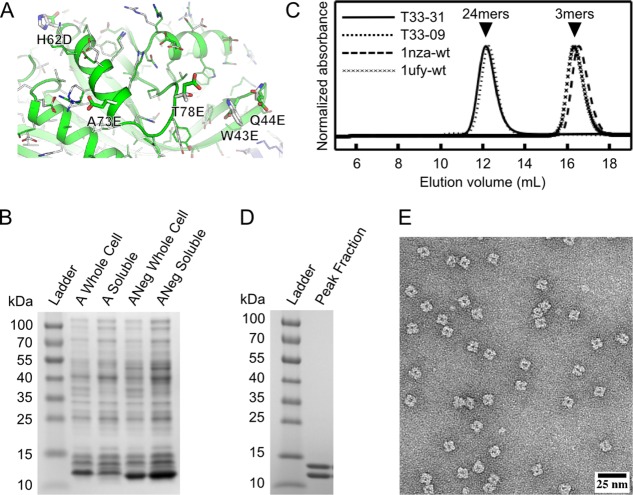
Figure 1.
Experimental characterization of designed protein assembly T33-31 by SDS-PAGE, analytical SEC, and electron microscopy. A: Close-up of the original subunit A and negatively charged subunit A (Aneg) from the T33-09 (white) and T33-31 (green and blue) design models. The five surface residues mutated in T33-31 compared to T33-09 are labeled and shown as sticks. B: SDS-PAGE analysis of whole cell and clarified lysates from cells expressing the original subunit A or the redesigned, negatively charged subunit A (ANeg). A strong band is observed near the expected molecular weight of 12.5 kDa in the clarified lysate of ANeg, but is only faintly visible in the subunit A sample. C: SEC chromatograms of purified designs and wild-type oligomeric proteins from which they are derived. The A and B subunits are derived from Protein Data Bank entries 1nza and 1ufy, respectively. The designed proteins elute near the expected volume for the target tetrahedral assembly (‘24mers’, arrow), while the wild-type proteins elute as trimers (‘3mers’, arrow). The T33-09 sample was produced from coexpressed subunits, whereas the T33-31 sample was produced through in vitro mixing as described in the Materials and Methods. D: SDS-PAGE analysis of SEC-purified T33-31. Two bands, with approximately equal intensity, are observed near the expected molecular weights of 12.5 and 14.5 kDa. E: Negative stain electron micrograph of in vitro-mixed, SEC-purified T33-31.