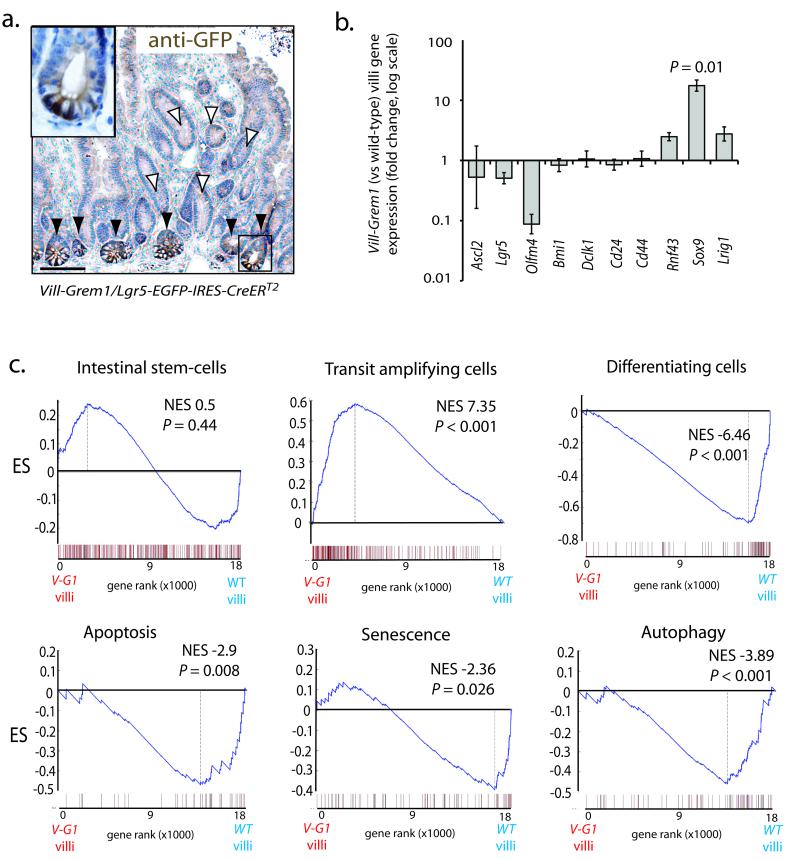
Fig. 3. Gene expression analysis of separated crypt/villus compartments in Vill-Grem1 mice.
(a) Vill-Grem1 mice were crossed with Lgr5-EGFP-IRES-CreERT2 mice and tissues stained with anti-GFP antibody. No Lgr5/EGFP positive cells were seen in the ectopic crypts on the villi (white arrowheads) but were found as expected in the underlying crypt bases (black arrowheads and inset). (b) qRT-PCR analysis of villus stem-cell marker expression in individual Vill-Grem1 villi versus wild-type littermate villi. There was a significant increase in expression of Sox9 (n = 10, P = 0.01, t-test) in Vill-Grem1 villi. (c) Villi from Vill-Grem1 and wild-type littermates underwent gene expression microarray and GSEA analysis using established gene program sets. GSEA plots shown are for Vill-Grem1 (n = 5) versus wildtype villi (n = 6). Enrichment score is calculated using Kolomogrov-Smirnov test (Supplementary reference8). P-value is calculated using a permutation test. Scale bars are 100 μm.