Fig. 4.
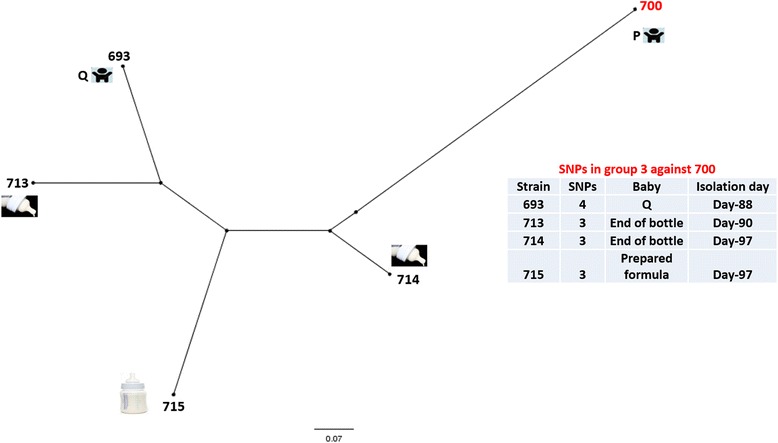
SNP phylogeny of the cluster strains. The SNPS were called using SMALT and SAMtools to generate the VCF files which were filtered using VCFTools to include only SNPs with minimum quality score of 30, minimum depth of 8, and minimum allele frequency of 0.90. The SNPs in the cluster were concatenated and used to create a maximum likelihood phylogeny. The red font colour indicates the reference strain. The tree was rooted to midpoint. The scale bar indicates the rate of nucleotide differences per sequence site
