Abstract
Background
Tyrosine kinase inhibitors (TKIs) are the recommended treatment for patients with chronic myeloid leukemia (CML). The MDR1/ABCB1 gene plays a role in resistance to a wide spectrum of drugs, including TKIs. However, the association of MDR1/ABCB1 gene polymorphisms (SNPs) such as C1236T, G2677T/A, and C3435T with the clinical therapeutic evolution of CML has been poorly studied. We investigated these gene polymorphisms in CML-patients treated with imatinib, nilotinib and/or dasatinib.
Methods
ABCB1-SNPs were studied in 22 CML-patients in the chronic phase (CP) and 2 CML-patients in blast crisis (BC), all of whom were treated with TKIs, and compared with 25 healthy controls using nested-PCR and sequencing techniques.
Results
Seventeen different haplotypes were identified: 7 only in controls, 6 only in CML-patients, and the remaining 4 in both groups. The distribution ratios of homozygous TT-variants present on each exon between controls and CML-patients were 2.9 for exon 12, and 0.32 for the other 2 exons. Heterozygous T-variants were observed in all controls (100%) and 75% of CML-patients. Wt-haplotype (CC-GG-CC) was observed in 6 CML-patients (25%). In this wt-group, two were treated with nilotinib and reached a major molecular response. The remaining 4 cases had either a minimal or null molecular response, or developed bone marrow aplasia.
Conclusion
Our results suggest that SNPs of the MDR1/ABCB1 gene could help to characterize the prognosis and the clinical-therapeutic evolution of CML-patients treated with TKIs. Wt-haplotype could be associated with a higher risk of developing CML, and a worse clinical-therapeutic evolution.
Keywords: CML, TKIs, ABCB1, SNPs, BCR-ABL
INTRODUCTION
Chronic myeloid leukemia (CML) is a myeloproliferative disease originating in hematopoietic stem-cells. It develops from the oncogenic activity of a 210kD protein (p210BCR-ABL), which is the product of a chimeric gene BCR-ABL1 with high activity tyrosine kinase (TK). One of the treatments of choice is based on the use of tyrosine kinase inhibitors (TKIs), such as imatinib, which blocks the ATP binding site in the catalytic pocket of the protein BCR-ABL1 [1,2,3]. Imatinib prevents the phosphorylation of proteins involved in different signaling pathways which promote cell proliferation and inhibit apoptosis [4,5]. Despite being a highly specific and effective molecular treatment for inhibiting TK, its therapeutic efficacy can be limited in some patients who develop a refractory phenotype at disease onset or during progression to both the accelerated phase and/or blast crisis [6,7,8]. An escalation in the dose of imatinib from 400 mg/d to 600-800 mg/d can be permitted in patients who had an initial suboptimal response or treatment failure, as defined by the European LeukemiaNet [9]. However these therapeutic schedules are not recommended for patients with imatinib intolerance, or who develop bone marrow aplasia as a secondary complication to imatinib and/or interferon treatment [10,11].
An alternative is to use other TKIs, such as dasatinib or nilotinib [3,12]. These are more commonly used in patients with mutations in the BCR-ABL gene which could generate failure of therapeutic response to imatinib. Another proposed TKI drug-resistance mechanism is that this can be generated by an insufficient drug concentration at the intracellular level, as the result of active transport being mediated by the over-expression of the ABC-transporters P-glycoprotein (P-gp) and/or BCRP (breast cancer resistant protein), encoded by the MDR1/ABCB1 and ABCG2 genes respectively [13,14]. Variations in single-nucleotide polymorphisms (SNPs) of MDR1/ABCB1 have been described as potential factors related to the clinical-therapeutic evolution of several diseases [15,16]. To date, 216 SNPs have been described in the ABCB1 gene (SNPer: http://snpper.chip.org), and several studies have identified polymorphic variants of TT in Exon 21 (T2677T) and in Exon 26 (T3435T) in Caucasian patients, which could also correlate with a phenotype of increased sensitivity to treatment with TKIs [17,18,19]. Other groups have described that ABCB1-SNP: C1236T on Exon 12 could also be related to a higher frequency of major molecular response in patients with CML treated with imatinib [20]. Taking together the above, it was suggested that association of the genotypes from these SNPs of the ABCB1 gene may configure a specific haplotype as a genetic biomarker of prognosis for therapeutic response in CML-patients treated with TKIs [21,22]. Here, we studied the SNPs C1236T, G2677T/A, and C3435T of the ABCB1 gene in CML-patients as potential predictive biological markers for therapeutic response and illness evolution.
MATERIALS AND METHODS
Patients and healthy controls
A total of 24 Caucasian patients with CML (Philadelphia chromosome-positive), with 22 cases in chronic-phase and the other 2 with blast crisis (BC), 11 females and 13 males aged between 21-76 years were included in the study. No patients with accelerated phase of the disease were enrolled. The end points were the rate of major molecular response at 6, 12 and 18 months after treatment started with imatinib (11 cases), nilotinib (6 cases), dasatinib (1 case), or combinations of 2 TKIs (6 cases). Cytogenetic and molecular evaluations were performed in accordance with international recommendations, and results were obtained from independent routine protocolized studies, performed during the clinical care and follow-up of the patients [23,24]. Blood samples from twenty-five healthy male volunteers were studied (age range, 22-54 years). For study of SNPs, DNA was obtained from peripheral blood samples with EDTA-tubes in all cases. The study was approved by the ethics committee of the School of Pharmacy and Biochemistry (University of Buenos Aires). All patients and healthy controls gave written informed consent. Previously to the development of this study, all enrolled cases were evaluated for the more common mutations of catalytic site of ABL-kinase, which had negative results.
DNA purification
Genomic DNA was extracted from peripheral blood leukocytes of both patients and controls using a commercial kit (Genomic DNA Purification kit, Thermo Scientific, Waltham, MA). Ultra PURE TRIzol reagent and molecular biology grade agarose were purchased from Gibco BRL (Life technologies, Paisley, UK), and were used according to the supplier's recommendations. DNA samples were stored at -70℃ until use.
PCR method
Each amplification reaction was performed with 5 µL (5 ng/mL) of genomic DNA in a 50 µL total reaction volume containing: Buffer Taq 10X (cc final 1X) 5 µL; DNTPs 2 mM (cc final 0.2 nM) 5 µL; MgCl2 50 mM (final concentration: 2.5 mM) 1.50 µL; Oligonucleotides Forward/Reverse 20 µM (final concentration: 200 nM) 0.25 µL; Taq polymerase 50 U/µ (Invitrogen) (final concentration: 0.025 U) 0.25 µL; H2O (DPC for PCR to final volume 40 µL) 38.00 µL; and ADN (sample patients) or H2O (negative control) 10 µL. The polymorphisms (SNPs) corresponding to the ABCB1 gene from exons 12 (C1236T; rs1128503), 21 (G2677T/A; rs2032582) and 26 (C3435T; rs1045642), were located in GenBank (NCBI Reference Sequence: NG_011513.1) accession CCDS5608 and classified according to each haplotype as: wild type homozygote, CC-GG-CC; heterozygote, T (or A) variant in at least one exon; full mutated homozygote, TT-TT/A-TT.
Primers for SNPs C1236T, G2677T/A and C3435T were designed according to previous descriptions [25,26,27] and the link: http://www.genatlas.org/. PCR was performed on genomic DNA using primer forward and reverse as follows: exon 12 forward, 5'-TCC-TGT-GTC-TGT-GAA-TTG-CCT-TG-3'; exon 12 reverse, 5'-GCT-GAT-CAC-CGC-AGT-CTA-GCT-CGC-3'; exon 21 forward, 5'-GTT-TTG-CAG-GCT-ATA-GGT-TCC-3'; exon 21 reverse, 5'-TTT-AGT-TTG-ACT-CAC-CTT-3'; exon 26 forward, 5'-GAT-CTG-TGA-ACT-CTT-GTT-TTC-A-3'; exon 26 reverse, 5'-GAA-GAG-AGA-CTT-ACA-TTA-GGC-3'.
The amplification program for exon 12 was: 95℃/5 min; 40 cycles of 95℃/30s, 55℃/30s, 72℃/60s; and 72℃/5 min for ending extension. The amplification program for exons 21 and 26 was: 95℃/5 min; 35 cycles at 95℃/45s, 55℃/45s, 72℃/60s, and 72℃/5 min for ending extension. Both programs were carried out in a GeneAmp PCR System 2400 (Roche Diagnostic Systems). The amplification products were analyzed on standard 1.5% agarose gels stained with ethidium bromide (0.5 µg/mL), and observed by UV transilluminator (ECX Compact; Vilber Lourmat; Belgium). All PCR products were purified and sequenced by automatic system ABI3730XL (Macrogen Inc., Korea). Exons 21 and 26 were studied in all controls (N=25) and all CML-patients (N=24), while exon 12 was studied in 19 controls and all CML-patients (N=24). Complete haplotype comparison was performed between 19 control and 24 CML-patients.
Statistical analysis
All the statistical analyses were performed with Statistical Package Medcalc 11.5 y VCCstat 2.0. For all variables, frequencies of distribution or percentage from total cases were established. All P values were analyzed by Mann Whitney or Fisher significance tests. The level of significance was taken as P<0.05. Signed consent in accordance with the Helsinki declaration was obtained from all patients as well as from healthy controls.
RESULTS
Twenty four CML-patients, 22 (91.7%) with chronic phase and 2 (8.3%) under blast crisis, were studied. Thirteen (54.2%) were males, and 11 (45.8%) were females. The Sokal risk was low in 7 (29.2%), intermediate in 7 (29.2%), high in 6 (25%), and not evaluated in 4 (16.6%). Age, gender, phase of disease, TKI treatment and doses, molecular responses (at 6, 12, and 18 months) and haplotypes are listed in Table 1.
Table 1. Characteristics and molecular responses of CML-patients.

a)Patients 7 and 13 had similar responses at both 6 and 18 months.
Abbreviations: CP, chronic phase; BC, blast crisis; BrCa, breast cancer; C, complete; Ma, major; Mi, minor; I, Imatinib; N, Nilotinib; D, Dasatinib; R, Responsive; NR, not-responsive; NE, not-evaluated; Int, intermediate; BMA, bone marrow aplasia.
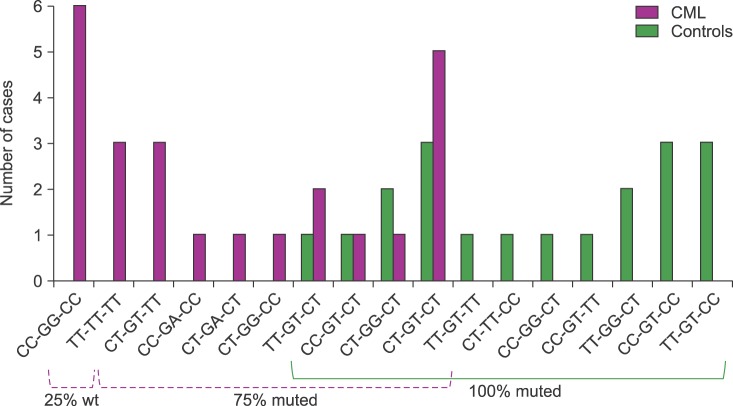
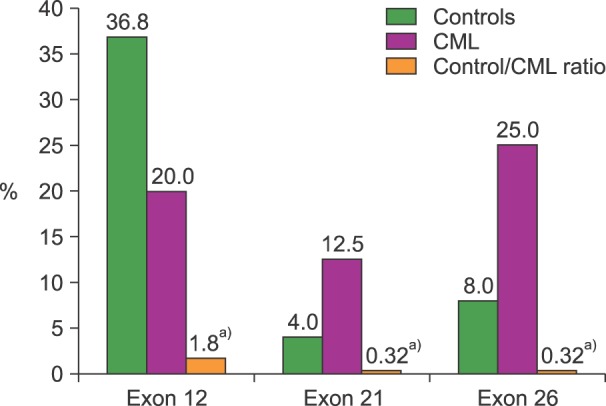
A total of seventeen different haplotypes were identified in CML-patients and controls. The frequency distribution indicates that 6 were only detected in CML-patients, 7 only in controls, and 4 in both groups. Heterozygous T-variant genotypes were observed in all 25 controls (100%), and in 18 CML-patients (75%). In this last group, 9 different T-variant haplotypes were identified as follows: full mutated-homozygote (TT-TT-TT) in 3 (12.5%), full mutated-heterozygous (CT-GT/A-CT) in 6 (25%), and other combinations of T-variants in 9 cases (37.5%). The remaining 6 (25%) CML-patients were wt-homozygous CC-GG-CC (Fig. 1). No associations between Sokal risk with haplotypes or therapeutic response were observed. TT-variant distributions for each exon indicate a predominant expression in exon 12 for controls but in exons 21 and 26 for CML-patients (Fig. 2).
Fig. 1. Frequency distribution of haplotypes in CML-patients and controls. CML were 25% wt and 75% muted, while controls were 100% muted.
Fig. 2. Frequency of TT-homozygous variants in each exon studied in controls and CML-patients. a)P<0.01, Mann-Whitney test.
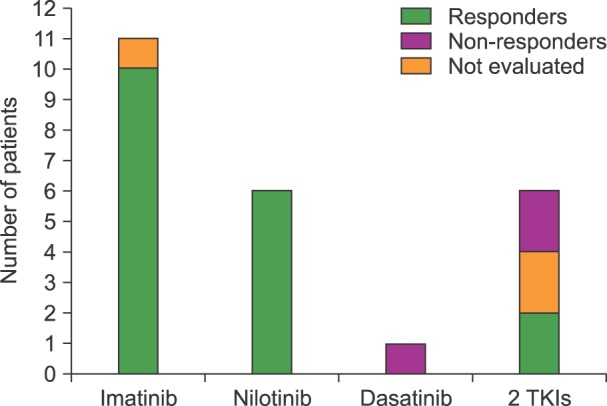
The therapeutic responses to different treatments were evaluated at the molecular level at three different end points (6, 12 and 18 months) (Table 1). However, for statistical comparison, only the end point at 12 month was evaluated. A total of 11 cases were treated with imatinib, 6 with nilotinib, and one with dasatinib as first and sole TKI; the other 6 cases were treated with the addition of dasatinib (4) or nilotinib (2) after initial administration of imatinib. Eleven patients received imatinib or imatinib+hydroxyurea (HU), and one of them who had a wt-haplotype (CC-GG-CC), developed bone marrow aplasia. This patient had a complex cytogenetic variant [46XX, t(3q2.5;9q3.4;22q1.1)] described only in a minority (2-10%) of CML-patients. The other ten cases also treated with imatinib were T-variants, and reached an MMR at six months. Similar responses were observed in the six patients (4 T-variants and 2 wt) treated with nilotinib. An earlier MMR response at 3 months was observed in the sole patient treated with dasatinib alone, however this patient relapsed with a minor molecular response (Mi) at 12 months; he also had wt-haplotype CC-GG-CC. The remaining 6 patients were treated with a combination of 2 TKIs as imatinib+dasatinib (N=4) or imatinib+nilotinib (N=2). One of these cases developed a breast cancer, another case in BC died within the first month, and another case that also had the wt-haplotype CC-GG-CC remained with a null response at 18 months (Fig. 3).
Fig. 3. Therapeutic responses of the 24 CML-patients at 12 months according to the TKI administered. Wt-haplotypes are indicated in each case. The remaining patients were T-variants. In three cases, therapeutic responses were not evaluated because the patients developed bone marrow aplasia, breast cancer, or early fatal blast crisis.
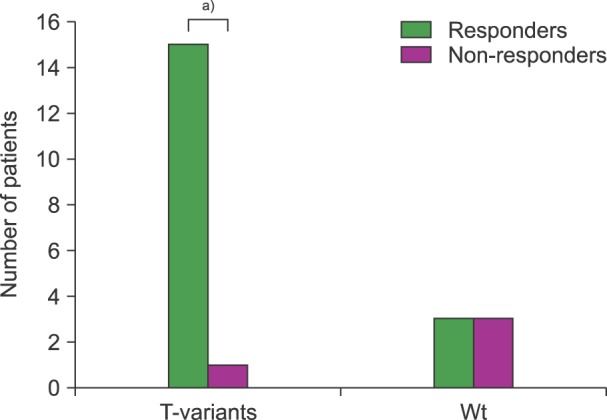
From a total of 24 CML-patients studied, 2 were in blast crisis at the time of diagnosis, and presented with haplotypes showing T-variants in all studied exons (CT-GT-TT and CT-GA-CT). This last case, who was the only patient with an allelic variant "A" in exon 21, was in BC and died during the first month after treatment was started. Excluding the BC cases, 16 of 22 CML-patients in the chronic phase were T-variants (heterozygotes and/or homozygotes). One of these cases developed a breast cancer, and so their therapeutic response was not evaluated. The remanding 15 patients were all responders (P<0.001). In the 6 cases with wt-haplotype only 3 (50%) were responders; of the others two had a null molecular response at 12 months and one developed bone marrow aplasia mentioned above (Fig. 4).
Fig. 4. Frequency of T-variants (with at least one T allele) in 22 CML-patients in the chronic phase according to their therapeutic response. a)P<0.001, X2-Fischer test.
DISCUSSION
CML affects mainly adults with a peak incidence between 46-55 years of age and a homogeneous distribution between genders, and in our study, the age and gender distribution was coincident with previous reports [3,28]. In our study, the frequency of heterozygous variants on exon 12 (C1236T) was similar to the previous description of Dulucq et al. [20]; however, we observed a higher percentage of these variants for exon 21 (2677G>/TA) (50% vs. 39.5%) and exon 26 (C3435T) (36.4% vs. 18.9%) [20]. Considering the relationship between allelic variants and therapeutic responses, 16/22 cases with CP were mutated (homozygous or heterozygous) and 15 of them (93.8%) reached a good molecular response at 6-18 months after treatment. The remaining case developed breast cancer and her therapeutic response was not evaluated. In the 6 cases in CP with full wt-homozygous haplotypes, 5 had an apparently good cytogenetic response; however, at the molecular level, 2 were refractory to treatment, and in another patient who developed bone marrow aplasia, the therapeutic response at the molecular level could not be evaluated. All this data reinforces the higher sensitivity of molecular methods for the measurement of minimal residual disease, according to the latest international recommendations for the management of CML [29,30].
To date, there are few studies of SNPs of MDR1/ABCB1 gene in patients with CML suggesting a potential relationship between genotype/haplotype studies and therapeutic responses when evaluated at the molecular level. Dulucq et al. [20] reported for the first time the association of mutated variants of exons 12 and 21 with an increased number of cases reaching a major molecular response (MMR). Additionally, they observed that wt-haplotypes (CC/GG/CC) had a lower frequency of MMR. Our results were in agreement with these observations. Maffioli et al. [21] state that the presence of a T allele only in exon 26 was associated with a better therapeutic response, while the haplotype characterized by the presence of a T allele in exon 12 and accompanied by mutant variants in exons 21 and 26 (1236T-2677G-3435C) was associated with primary resistance to imatinib. A different conclusion was recently reached by Angelini et al. [22], indicating that the wt genotype (CC) in exon 26 was associated with a complete molecular response, while T-variant haplotypes in all three exons did not have any relation with therapeutic responses. However, our study appears to indicate a better therapeutic response in patients with T-variants present in all these exons, because 93.75% of them reached MMR at 18 months. On the other hand, we observed a potential association between the genotypes and/or haplotypes detected with the risk of developing the disease (Fig. 1 and 2). In this regard, our study demonstrated that the full wt-haplotype CC-GG-CC (6/24 cases) was only present in patients with CML, and this haplotype was related in 3 of them with poor prognoses showing minor (one case) or null (one cases) responses at 18 months, and developing bone marrow aplasia in one case. Interestingly one other case had a null response during the first 12 months, but reached an MMR at 18 months.
In conclusion, we suggest that combined evaluation of haplotypes and genotypes of the SNPs of the MDR1/ABCB1 gene on exons 12, 21 and 26 could be a useful predictive tool for both therapeutic response as well as the risk of developing and the clinical evolution of CML.
ACKNOWLEDGMENTS
We thank Lic Nora Castiglia for her help in the statistical analyses.
Footnotes
This study was supported by UBACyT (CB04).
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
References
- 1.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 2.Saglio G, Kim DW, Issaragrisil S, et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. N Engl J Med. 2010;362:2251–2259. doi: 10.1056/NEJMoa0912614. [DOI] [PubMed] [Google Scholar]
- 3.Goldman JM, Melo JV. Chronic myeloid leukemia-advances in biology and new approaches to treatment. N Engl J Med. 2003;349:1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- 4.Nagar B, Bornmann WG, Pellicena P, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 5.Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 6.Druker BJ, Guilhot F, O'Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 7.Hochhaus A, Kreil S, Corbin AS, et al. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002;16:2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 8.Deininger MW, Druker BJ. SRCircumventing imatinib resistance. Cancer Cell. 2004;6:108–110. doi: 10.1016/j.ccr.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Baccarani M, Saglio G, Goldman J, et al. Evolving concepts in the management of chronic myeloid leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2006;108:1809–1820. doi: 10.1182/blood-2006-02-005686. [DOI] [PubMed] [Google Scholar]
- 10.Khan KA, Junaid A, Siddiqui NS, Mukhtar K, Siddiqui S. Imatinib-related bone marrow aplasia after complete cytogenetic response in chronic myeloid leukemia. J Coll Physicians Surg Pak. 2008;18:176–178. [PubMed] [Google Scholar]
- 11.Alabdulaaly A, Rifkind J, Solow H, Messner HA, Lipton JH. Rescue of interferon induced bone marrow aplasia in a patient with chronic myeloid leukemia by allogeneic bone marrow transplant. Leuk Lymphoma. 2004;45:175–177. doi: 10.1080/1042819031000139774. [DOI] [PubMed] [Google Scholar]
- 12.Henkes M, van der Kuip H, Aulitzky WE. Therapeutic options for chronic myeloid leukemia: focus on imatinib (Glivec, Gleevectrade mark) Ther Clin Risk Manag. 2008;4:163–187. [PMC free article] [PubMed] [Google Scholar]
- 13.Mahon FX, Hayette S, Lagarde V, et al. Evidence that resistance to nilotinib may be due to BCR-ABL, Pgp, or Src kinase overexpression. Cancer Res. 2008;68:9809–9816. doi: 10.1158/0008-5472.CAN-08-1008. [DOI] [PubMed] [Google Scholar]
- 14.Shukla S, Sauna ZE, Ambudkar SV. Evidence for the interaction of imatinib at the transport-substrate site(s) of the multidrug-resistance-linked ABC drug transporters ABCB1 (P-glycoprotein) and ABCG2. Leukemia. 2008;22:445–447. doi: 10.1038/sj.leu.2404897. [DOI] [PubMed] [Google Scholar]
- 15.Tanabe M, Ieiri I, Nagata N, et al. Expression of P-glycoprotein in human placenta: relation to genetic polymorphism of the multidrug resistance (MDR)-1 gene. J Pharmacol Exp Ther. 2001;297:1137–1143. [PubMed] [Google Scholar]
- 16.Gréen H, Söderkvist P, Rosenberg P, Horvath G, Peterson C. mdr-1 single nucleotide polymorphisms in ovarian cancer tissue: G2677T/A correlates with response to paclitaxel chemotherapy. Clin Cancer Res. 2006;12:854–859. doi: 10.1158/1078-0432.CCR-05-0950. [DOI] [PubMed] [Google Scholar]
- 17.Marzolini C, Paus E, Buclin T, Kim RB. Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin Pharmacol Ther. 2004;75:13–33. doi: 10.1016/j.clpt.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Riva A, Kohane IS. SNPper: retrieval and analysis of human SNPs. Bioinformatics. 2002;18:1681–1685. doi: 10.1093/bioinformatics/18.12.1681. [DOI] [PubMed] [Google Scholar]
- 19.Jamroziak K, Robak T. Pharmacogenomics of MDR1/ABCB1 gene: the influence on risk and clinical outcome of haematological malignancies. Hematology. 2004;9:91–105. doi: 10.1080/10245330310001638974. [DOI] [PubMed] [Google Scholar]
- 20.Dulucq S, Bouchet S, Turcq B, et al. Multidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood. 2008;112:2024–2027. doi: 10.1182/blood-2008-03-147744. [DOI] [PubMed] [Google Scholar]
- 21.Maffioli M, Camós M, Gaya A, et al. Correlation between genetic polymorphisms of the hOCT1 and MDR1 genes and the response to imatinib in patients newly diagnosed with chronic-phase chronic myeloid leukemia. Leuk Res. 2011;35:1014–1019. doi: 10.1016/j.leukres.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Angelini S, Soverini S, Ravegnini G, et al. Association between imatinib transporters and metabolizing enzymes genotype and response in newly diagnosed chronic myeloid leukemia patients receiving imatinib therapy. Haematologica. 2013;98:193–200. doi: 10.3324/haematol.2012.066480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. WHO Classification of tumours. pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon, France: IARC Press; 2001. pp. 20–26. [Google Scholar]
- 24.Baccarani M, Cortes J, Pane F, et al. Chronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27:6041–6051. doi: 10.1200/JCO.2009.25.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni LN, Li JY, Miao KR, et al. Multidrug resistance gene (MDR1) polymorphisms correlate with imatinib response in chronic myeloid leukemia. Med Oncol. 2011;28:265–269. doi: 10.1007/s12032-010-9456-9. [DOI] [PubMed] [Google Scholar]
- 26.Illmer T, Schuler US, Thiede C, et al. MDR1 gene polymorphisms affect therapy outcome in acute myeloid leukemia patients. Cancer Res. 2002;62:4955–4962. [PubMed] [Google Scholar]
- 27.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger U, Maywald O, Pfirrmann M, et al. Gender aspects in chronic myeloid leukemia: long-term results from randomized studies. Leukemia. 2005;19:984–989. doi: 10.1038/sj.leu.2403756. [DOI] [PubMed] [Google Scholar]
- 29.Baccarani M, Deininger MW, Rosti G, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122:872–884. doi: 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahon FX, Etienne G. Deep molecular response in chronic myeloid leukemia: the new goal of therapy? Clin Cancer Res. 2014;20:310–322. doi: 10.1158/1078-0432.CCR-13-1988. [DOI] [PubMed] [Google Scholar]