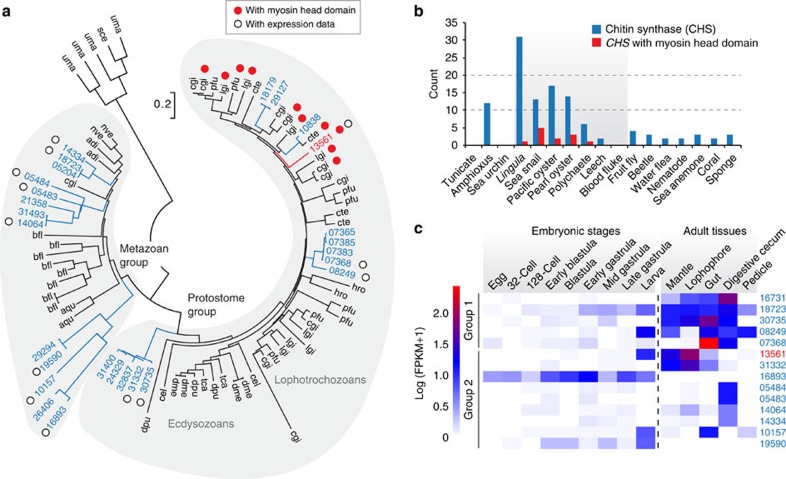
Figure 3. Expansion and expression of Lingula chitin synthase genes indicate roles in shell formation and digestion.
(a) Phylogenetic analysis of chitin synthase (CHS) genes using the neighbour-joining method with the JTT model (90 genes, 358 amino acids and 1,000 bootstrap replicates). Three-letter code: adi, coral (Acropora digitifera); aqu, sponge (Amphimedon queenslandica); bfl, amphioxus (B. floridae); cel, nematode (Caenorhabditis elegans); cgi, Pacific oyster (Cr. gigas); cte, polychaete (Ca. teleta); dme, fly (Drosophila melanogaster); dpu, water flea (Daphnia pulex); hro, leech (H. robusta); lgi, sea snail (L. gigantea); nve, sea anemone (Nematostella vectensis); pfu, pearl oyster (Pinctada fucata); sce, baker's yeast (Saccharomyces cerevisiae); tca, beetle (Tribolium castaneum); uma, corn smut fungus (Ustilago maydis). Numbers are Lingula gene IDs. (b) CHS genes detected with BLASTP among 17 selected metazoan genomes. It is noteworthy that CHS genes with myosin head domains are only present among lophotrochozoans (grey area). (c) The expression of Lingula CHS genes in embryonic stages and adult tissues (separated by a vertical dashed line). FPKM, fragments per kilobase of transcript per million mapped reads.