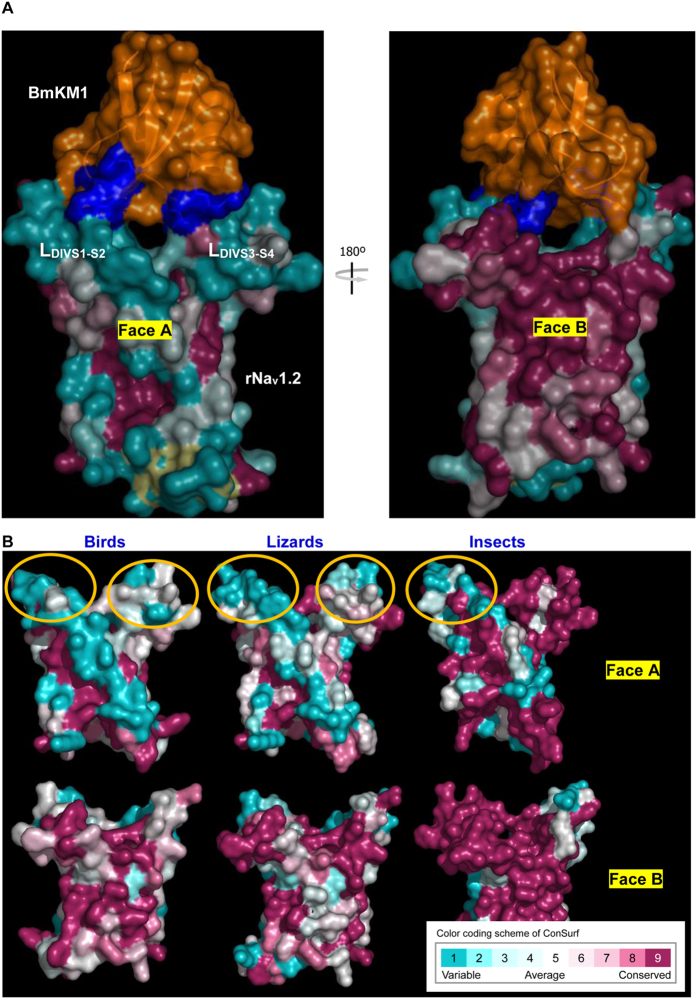
Figure 7. High variability of scorpion α-toxin-bound regions in Nav channels from both predators and prey of scorpions.
(A) Molecular surface display shows that BmKM1 binds to the evolutionarily variable loops (LDIVS1-S2 and LDIVS3-S4) of the VSD via its PSSs (blue). The complex model is the same with that of Fig. 3; (B) Molecular surfaces of VSDs from birds, lizards and insects. Variable regions involved in toxin binding are circled. Consurf (http://consurf.tau.ac.il/) was introduced to compute the position-specific conservation scores in the VSD and colored according to the scores. Structures of bird, lizard and insect VSDs are modeled from sequences of Picoides pubescens (gi|699624821), Anolis carolinensis (gi|343098400) and Drosophila melanogaster (gi|403447) (Figs. S2, S3 and S5) based on the previously reported rNav1.2 model29. Faces A and B are two faces of the VSD rotated 180° around y axis.