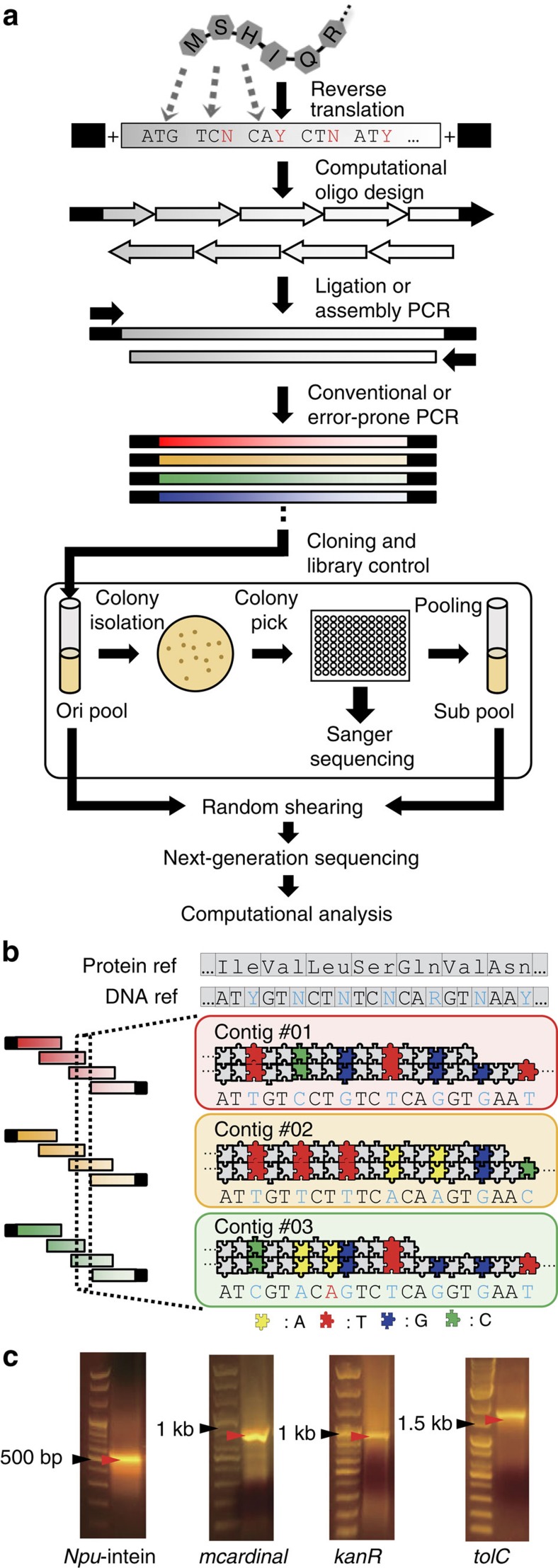
Figure 1. Overview of JigsawSeq pipeline.
(a) General scheme of gene library construction leading to NGS. Native protein sequences were reverse-translated with barcodes (N, R and Y). Forward and reverse flanking sequence were attached to both ends of reverse-translated DNA sequences. Black regions represent flanking vector sequences. After LCR or assembly PCR, assembled genes were cloned into an appropriate vector. Sub-pools (Sub) were generated by isolating 96 colonies from its initial pool. Then, these pools were subjected to standard NGS library preparation. For performance validation, we verified 96 colonies by Sanger sequencing. (b) Schematic representation of overview of de Bruijn graph-based assembly of a codon-barcoded library. Codon-barcoded reads interlock with each other to assemble final contigs. (c) Library insert sizes were verified by agarose gel electrophoresis.