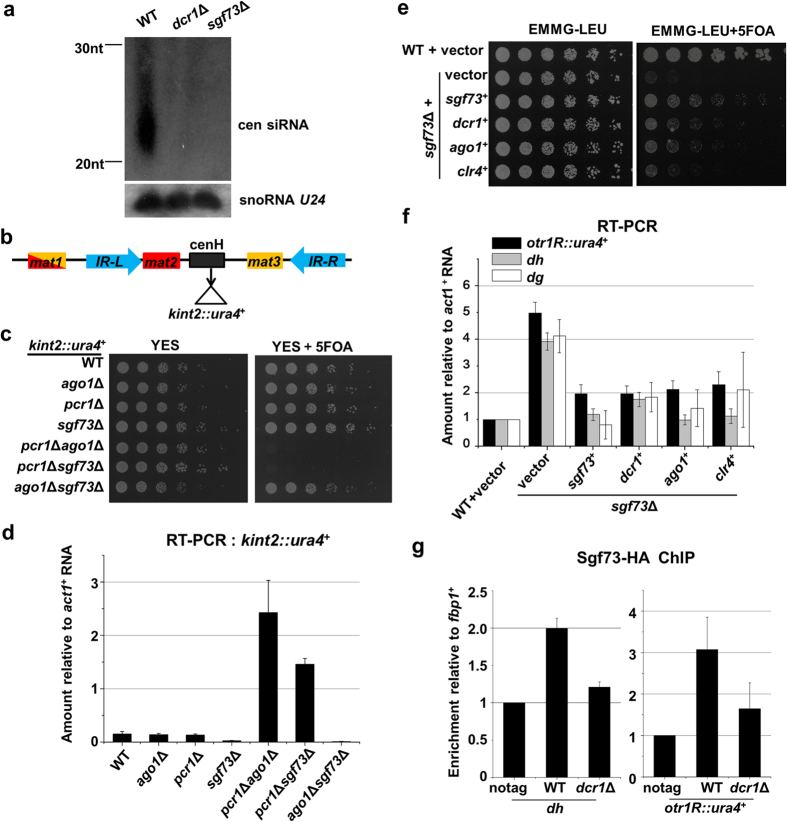
Figure 3. Sgf73 is an essential component of RNAi machinery.
(a) Northern blot analysis of centromeric siRNAs using probes against dg and dh. snoRNA U24 was detected as a loading control. (b) A schematic representation of mating type region and the position of a marker gene inserted into K region (kint2::ura4+). (c) Five-fold serial dilution assay to examine the silencing of kint2::ura4+ in sgf73Δ cells, and in strains combining deletion of Sgf73 and deletion of a effector in the RNAi or Atf1/Pcr1 pathway. (d) RT-PCR analysis of kint2::ura4+ RNA levels from the strains in (c). The relative level to a control act1+ in WT cells was arbitrarily designated as 1 Each column shown in (d) and below represents the mean ± s.d. from three biological repeats. (e) Fivefold serial dilution assay to examine the silencing of otr1R::ura4+ in sgf73Δ cells overexpressing dcr1+, ago1+ or clr4+. sgf73Δ cells were transformed with a plasmid overexpressing the indicated gene and transformants were subject to the silencing assay. (f) RT-PCR analysis of dh, dg and otr1R::ura4+ RNA levels relative to a control act1+ in the transformants in (e). (g) ChIP analysis of enrichment of Sgf73-3HA at dh and otr1R::ura4+ relative to fbp1+. Relative enrichment in the cells without tagging (no tag) was arbitrarily designated as 1.