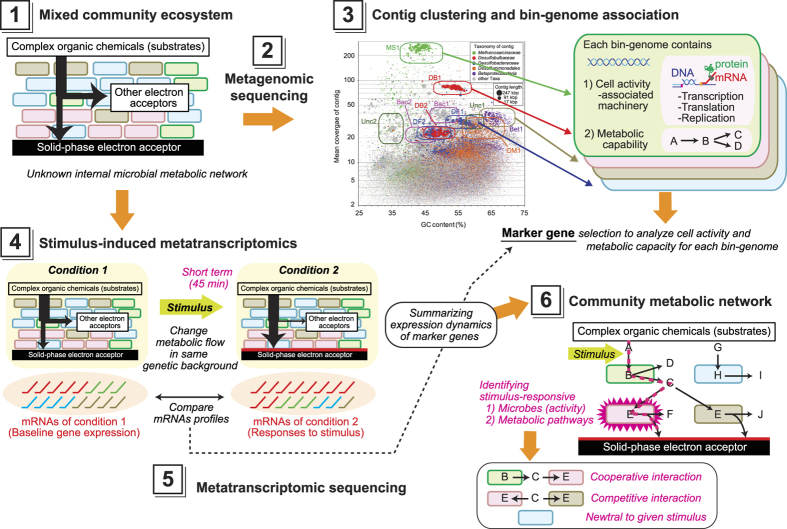
Figure 1. Scheme for the analytical approach used to describe the microbial networks in a complex EET-active community.
(Step 1) Enrichment of an EET-active microbial ecosystem; (Step 2) Metagenomic sequencing and analysis of the community; (Step 3) Bin-genome association by contig clustering. Each cluster indicates a Bin-genome of a community member, which includes coding sequences for cell activity and available metabolic pathways; (Step 4) stimulus-induced metatranscriptomics involving the application of a specific EET-condition via stimulus addition and biofilm sampling with DNA and mRNA extraction; (Step 5) Metatranscriptomic sequencing and subsequent comparative analyses of gene expression profiles of the whole community and each community member, which are executed via marker gene sets correlated with important “cell activity” and “metabolic” functions; (Step 6) Construction of the community metabolic network for the dominant microbes within the community.