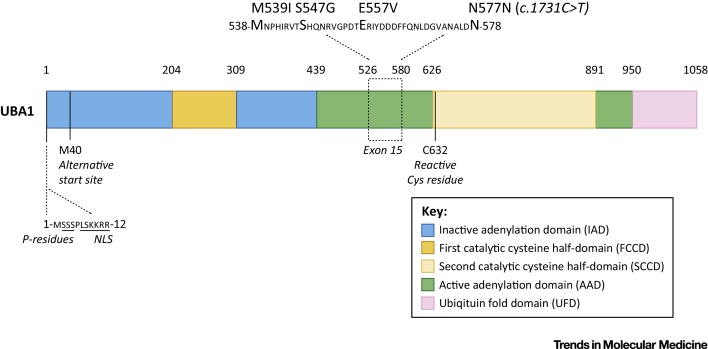
Figure 1.
Domains of Ubiquitin-Like Modifier Activating Enzyme 1 (UBA1) and Genetic Variants Identified in X-Linked Spinal Muscular Atrophy (XL-SMA). The N-terminal half of UBA1 comprises an inactive adenylation domain (IAD) that surrounds the first catalytic cysteine half-domain (FCCD). The C-terminal half of UBA1 comprises an active adenylation domain (AAD) that surrounds the second catalytic cysteine half-domain (SCCD). The SCCD contains the reactive Cys residue that binds ubiquitin. The C-terminal ubiquitin fold domain (UFD) allows UBA1 to bind to E2 enzymes. When UBA1 is folded into its 3D structure, the FCCH and SCCH and the IAD and AAD are directly adjacent to each other [78,79] (Box 1). The Met residue at position 40 provides an alternative translational start site that leads to the expression of the UBA1b isoform of the protein. The UBA1a isoform-specific N-terminal sequence contains a nuclear localization signal (NLS) and Ser residues that can be phosphorylated (P residues). Mutations in UBA1 that have been shown to cause XL-SMA cluster in exon 15 of the protein. The domain structure in this figure is based on yeast [78] and mouse [79] UBA1 structural analysis. The specific amino acid positions in the figure are based on the mouse UBA1 sequence, as the mouse and human UBA1 protein sequences are more than 95% identical. The amino acids that are mutated in XL-SMA as well as the amino acid sequences that surround the borders of the various domains are all perfectly conserved between mouse and human UBA1 sequences.