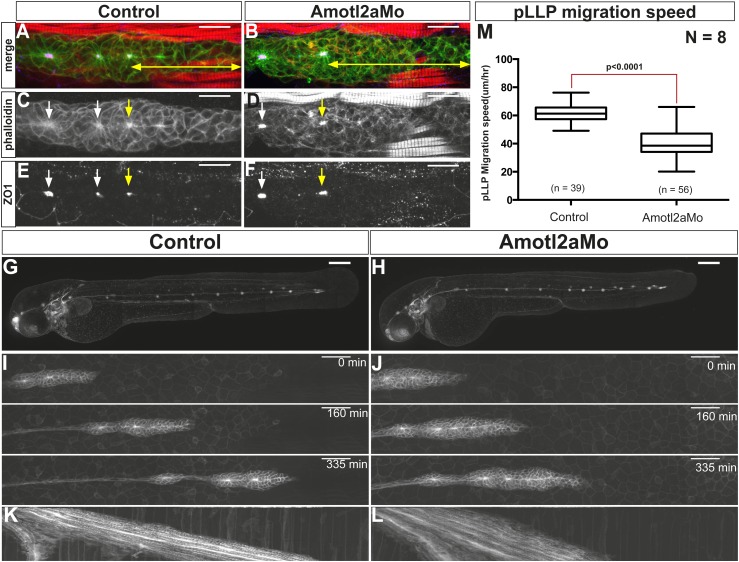
Figure 2. Amotl2a is not essential for proneuromast assembly but for proper migration.
(A–F) MIP of Z-stacks of pLLP stained with ZO-1 (blue) and GFP (green) antibodies and phalloidin (red) in control (A, C, E) and amotl2a morphant (B, D, F) cldnb:gfp embryos at 30 hpf. (G, H) MIP of overview images of control (G) and amotl2a morphant (H) embryos after completion of migration. (I, J) Snapshots of time-lapse movies at the indicated timepoints showing a delay in migration in amotl2a morphants (J) as compared to controls (I). (K, L) Corresponding kymographs used to measure the migration speed. (M) Boxplot comparing the migration speeds (Figure 2—source data 1, Figure 2—figure supplements 1, 2).
DOI: http://dx.doi.org/10.7554/eLife.08201.006