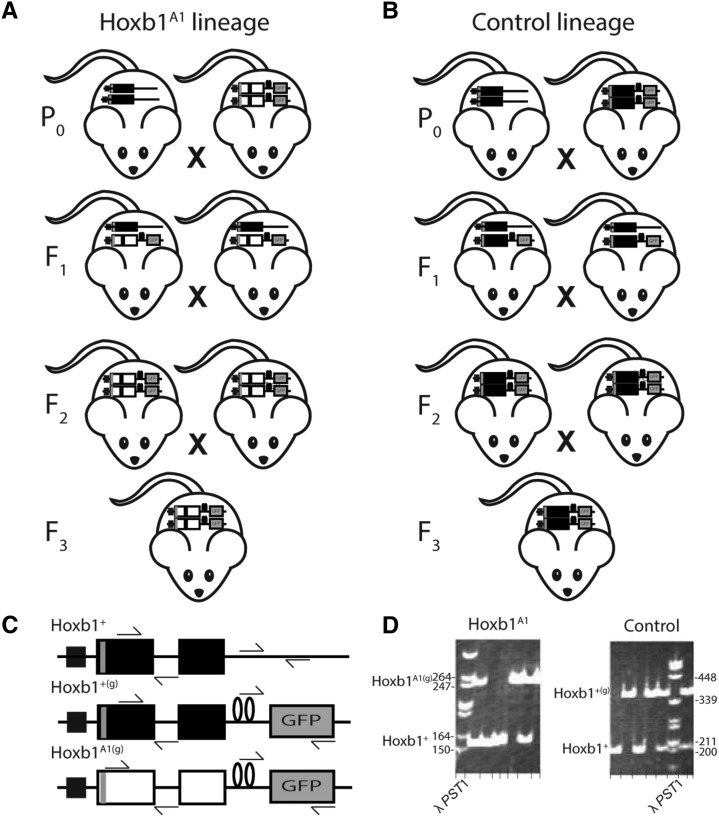
Figure 1.
Breeding design for production of Hoxb1A1 and control founders. (A) To produce animals bearing Hoxb1A1swaps that also possess the functional behaviors needed for OPAs, Hoxb1A1(g)/A1(g) 129 × C57BL/6 mice were bred to outbred, wild-derived mice. The resulting Hoxb1A1(g)/+ heterozygotes were crossed to establish the next (F2) generation. Progeny were genetically screened and only Hoxb1A1(g)/A1(g) individuals were selected as OPA founders. Three OPA populations were established with these F2 animals and three more were established with F3 animals produced from F2 Hoxb1A1(g)/A1(g) homozygous breeding pairs. (B) To control for potential confounding effects due to differential genetics surrounding the swap control animals were bred by crossing Hoxb1+(g)/+(g) 129 × C57BL/6 mice with the same wild stock used in the Hoxb1A1 treatment lineage. The Hoxb1+(g)/+ were then crossed to produce the F2 generation. Only Hoxb1+(g)/+(g) mice were selected as OPA founders. Three OPA populations were established with these F2 animals and three more were established with F3 animals produced from F2 Hoxb1+(g)/+(g) homozygous breeders. (C) Illustrations of wild type (Hoxb1+), wild type with the IRES–τ-GFP tag (Hoxb1+(g)), and Hoxb1A1 swap with the IRES–τ-GFP tag (Hoxb1A1(g)) are provided. Large rectangles represent exons 1 and 2 of Hoxb1 (black) and Hoxa1 (white). The Hoxb1 promoter is conserved across all genotypes and solid squares represent the Hoxb1 autoregulatory enhancer. Loops separating the τ-GFP tag from the second exon depict the IRES. Arrows approximate primer binding sites. Illustrations are not to scale. (D) Image of polyacrylamide gel discrimination between Hoxb1A1(g) and Hoxb1+ (left) and between Hoxb1+(g) and Hoxb1+(right) alleles.