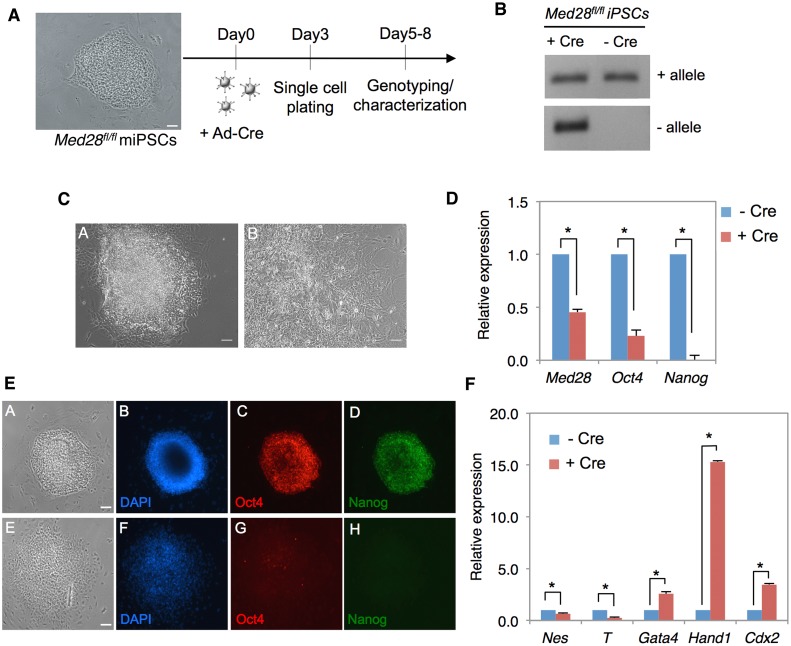
Fig 6. Cre-mediated heterozygous deletion of Med28 causes differentiation of iPSC.
(A) Med28 fl/fl iPSCs were infected with Ad-Cre, followed by single cell plating on day 3 and genotyping along with other analyses performed between days 5–8. (B) PCR genotyping shows clones with both deleted (- allele) and non-deleted WT (+ allele) alleles infected with Ad-Cre (+ Cre) compared to uninfected clones (-Cre), respectively. (C) Representative bright field images show noticeable differences in morphology between non-deleted (panel A, -Cre) and deleted (panel B, +Cre) Med28 fl/fl iPSC colonies (scale bar 200μm). Note the differentiated colony morphology of the Med28 fl/fl (panel B, +Cre) cells. (D) Quantitation of real-time PCR for deleted Med28 fl/fl iPSC colonies (+Cre) show ~2-fold decreased expression of Med28 as well as significantly lower expression of Oct4 and Nanog (n = 4; *, p<0.05) compared to non-deleted Med28 fl/fl iPSC (-Cre) colonies. (E) Immunostaining of deleted Med28 fl/fl colonies (panels E-H) show loss of self-renewal factor Oct4 (red, C and G) and Nanog (green, D and H) compared to non-deleted colonies (panels A-D). Bright field images (A and E) and nuclear DAPI (blue, B and F) are also shown (scale bar 200μm). (F) Quantitation of real—time PCR for differentiation markers reveal increased differentiation markers in deleted (+Cre) Med28 fl/fl iPSC colonies for extra embryonic lineage (Gata4, Hand1, and Cdx2) and decreased germ layer lineage makers nestin (Nes, ectoderm) and T brachyury (T, mesoderm) compared to non-deleted (-Cre) Med28 fl/fl iPSCs colonies (n = 4). Data are presented as mean +/- SD (*p<0.05).