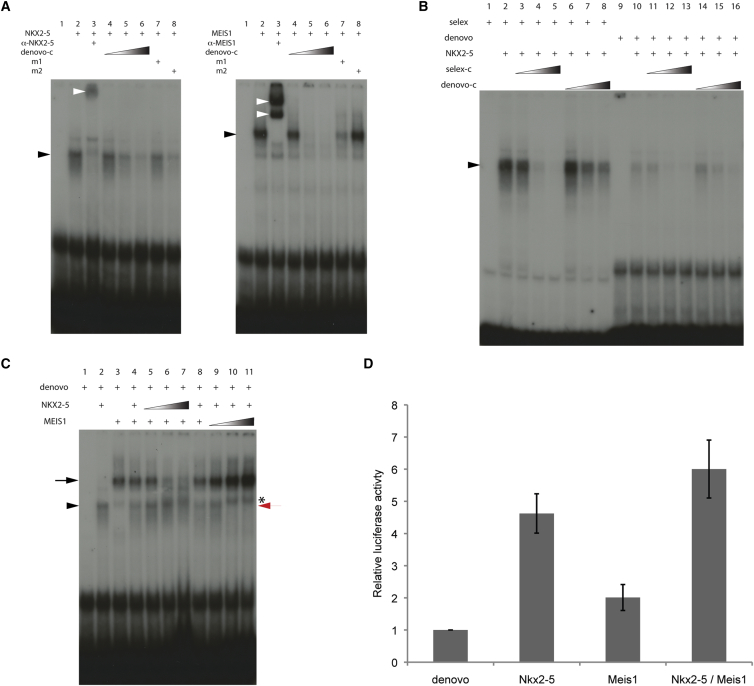
Figure 3.
NKX2-5 and MEIS1 Bind on the Same DNA Motif
(A) Electrophoretic mobility shift assay (EMSA) showing that NKX2-5 and MEIS1 bind the de novo probe (black arrowheads). Note the specific competition when an unlabelled competitor (de novo-c) is used. White arrowheads indicate supershift when a specific antibody against the protein tested is used. Note that competition with a mutated unlabelled probe is observed with de novo m2, but not de novo m1, when NKX2-5 protein is used. Competition with a mutated unlabelled probe is observed with de novo m1, but not m2, when MEIS1 protein is used.
(B) The selex unlabelled probe is a better competitor than the de novo unlabelled probe when used with selex or de novo radioactive probes. Black arrowhead indicates NKX2-5 probe complex.
(C) EMSA showing the absence of any supershift when both NKX2-5 and MEIS1 proteins are added in increasing amounts. Increasingly prominent band slightly above the position of the MEIS1 complex in lanes 10 and 11 (indicated with ∗) is detectable in the presence of MEIS1 alone in lane 3, and its apparent increase simply reflects the increase in overall MEIS1 concentration in the titration experiment. Black arrow and black arrowhead indicate MEIS1 and NKX2-5 binding on the de novo probe, respectively. Note the disappearance of NKX2-5 binding in lanes 10 and 11 at higher concentrations of MEIS1 (red arrow).
(D) NKX2-5 and MEIS1 activate the de novo fragment cloned in pGL3 in 3T3 cells. Note the absence of synergistic activation. The graph shows relative luciferase reporter activity normalized to reporter construct alone. Average of three independent experiments is shown. Error bars represent the SEM.