To understand the historical circumstances that shape populations of sympatric and closely related taxa, microsatellite markers were used, while populations of three Salvia species served as a study model. In the widespread S. officinalis no population genetic disturbances were detected, in the endemic S. brachyodon evidence for clonality and a genetic bottleneck were found while the results of the S. fruticosa population indicated high inbreeding levels and hybridization with S. officinalis. As many findings regarding demography of individual population or species can be reached only through their comparison with closely related taxa, this study demonstrates the importance of the multi-species approach.
Keywords: Hybridization, Mediterranean, population bottleneck, population genetics, Salvia, SSR
Abstract
Gene flow, natural selection and genetic drift are processes that play a major role in shaping the genetic structure of natural populations. In addition, genetic structures of individual populations are strongly correlated with their geographical position within the species distribution area. The highest levels of genetic variation are usually found in the centre of a species' distribution and tend to decrease beyond that point. Additionally, narrowly endemic taxa are expected to be characterized by lower levels of genetic variation than their widespread congeners. To understand the historical circumstances that shape populations of sympatric and closely related taxa, microsatellite markers were used, while populations of the three closely related and sympatric Mediterranean Salvia species (S. officinalis L., S. fruticosa Mill. and S. brachyodon Vandas) served as a study model. In the populations of widespread S. officinalis, located in the central parts of this species' distribution area, no population genetic disturbances were detected. The narrow endemic S. brachyodon showed heterozygote excess, clonal reproduction and a genetic bottleneck. Because the genetic bottleneck was likely caused by the disappearance of suitable open-type habitats, the recent wildfire that cleared the terrain probably saved the S. brachyodon population from gradual deterioration and extinction. At the same time, clonal reproduction could serve as a valuable mechanism in the preservation of genetic variability. The results of the disjunct S. fruticosa population indicated heterozygote deficiency, inbreeding, hybridization with S. officinalis and population expansion. The hybridization with S. officinalis along with the abandonment of the agro-pastoral system are likely the main drivers of the strong expansion of S. fruticosa in the studied location. As many relevant findings and conclusions regarding historical and contemporary demography of individual populations or species can be reached only through their comparison with closely related taxa, this study demonstrates the importance and advantages of such a multi-species approach.
Introduction
There are three processes that are believed to play major roles in shaping the genetic structure of natural populations: gene flow, natural selection and genetic drift (Lesica and Allendorf 1995; Eckert et al. 2008). Numerous past events, either natural or human-mediated, can have substantial effects on the current intra- and interspecific genetic variability. Interspecific hybridization, genetic bottlenecks, the founder effect, inbreeding depression and rapid population expansion are just some of these effects. Genetic drift events, such as the founder effect and bottlenecks, often lead to increased rates of inbreeding, a loss of heterozygosity and the fixation of deleterious alleles (Luikart et al. 1998). With the increased probability of the homozygous expression of recessive deleterious alleles, the risk of extinction increases for many species undergoing population bottlenecks (Luikart et al. 1998). Therefore, the early detection of a bottleneck event is of central interest to conservation biologists. Among other mechanisms, hybridization plays an important role in the evolution of many taxonomic groups and can result in the formation of entirely new species. Hybridization can be of natural or anthropogenic origin; it occurs with or without introgression and can also lead to adaptive evolution and diversification via heterosis or the production of novel genotypes capable of expressing superior phenotypic traits (Stebbins 1959; Arnold 1997; Rieseberg et al. 2003; Seehausen 2004).
Differences and contrasting patterns in genetic structures across core, edge and disjunct populations are the source of never ending debates and research (e.g. Gaston 2003; Sagarin et al. 2006; Samis and Eckert 2007; Eckert et al. 2008). It is generally assumed that the highest abundance and the highest levels of genetic variability can be found in the geographical centre of species distribution (Surina et al. 2014). As we move away from the species distribution centre, population sizes gradually decrease as does genetic diversity. Similarly, a disjunct population is expected to be characterized by impoverished genetic structure as a direct consequence of its origin associated with some sort of bottleneck event followed by the absence of inter-population gene flow (Meeus et al. 2012). The origin of disjunct populations is usually linked to one of three possible causes: (i) human introduction, (ii) long distance dispersal and (iii) isolation after species range contractions due to either climatologically or anthropogenically induced reasons (Meeus et al. 2012).
The Mediterranean basin is known not only for its glacial refugium areas that have played a crucial role in postglacial recolonizations of Europe (e.g. Taberlet et al. 1998; Hewitt 1999, 2001; Michaux et al. 2003) but also for the long-lasting human presence and the influence of humans on local ecosystems that can be dated back as far as 50 000 years ago (Naveh and Dan 1973). At the beginning of the Holocene, some 10 000 years ago, this anthropogenic pressure on living systems became more intense as plants and animals were domesticated, and gradually, a sustainable agro-pastoral system developed (Miller 1992; Zeder and Hesse 2000; Blondel 2006). Consequently, the vast majority of present-day Mediterranean landscapes and accompanying species communities serve as examples of ‘coevolution’ between nature and humans that have lasted since the last glacial period. During this time, human activities and practices have shaped the complex interaction between human societies and the local ecosystems and have subsequently contributed to maintaining high levels of biological diversity (Blondel 2006; Fonderflick et al. 2010).
With over 900 species distributed worldwide, the genus Salvia is by far the largest and most diverse genus in the Lamiaceae family (Walker et al. 2004). In Europe, 36 taxa are described and grouped into seven sections (Hedge 1972). Among them is the section Salvia, which comprises 13 taxa at species rank (Hedge 1972), of which Salvia officinalis L. (common sage) (Fig. 1) and S. fruticosa Mill. (Greek sage) (Fig. 1) are best known because of their economically relevant high proportions and quality of essential oil (Putievsky et al. 1990; Dudai et al. 1999). As a typical member of indigenous flora, S. officinalis naturally grows along the eastern Adriatic coast (Pignatti 1982), in the central and southern Apennines (Lucchese et al. 1995; Cutini et al. 2007; Di Pietro 2011), and a few relict populations can be found in continental parts of the Balkan Peninsula (Vasić 1997). The range of S. fruticosa extends from northeastern Libya to Sicily and southern Italy through the southern part of the Balkan Peninsula to western Syria (Hedge 1982; Karousou et al. 2000), but this species has been introduced to many Mediterranean countries throughout history (i.e. Malta, Spain, Portugal) by the Phoenicians and the Greeks (Karousou et al. 2000). In the central Adriatic region, an isolated population of Greek sage can be found on the Island of Vis (Croatia) (Flora Croatica Database; http://hirc.botanic.hr/fcd). Because this population is located ∼500 km from the nearest population of the same species in southern parts of the Republic of Albania, its origin is unclear, and it is uncertain whether this population is indigenous or naturalized, especially considering that in the 4th century BC, a well-organized Greek colony called Issa was founded at this location.
Figure 1.

(A) Salvia officinalis in full bloom in the Island of Vis, (B) S. fruticosa in abandoned olive groves and vineyards outside the local settlement in the Island of Vis, (C) S. brachyodon in Pelješac Peninsula, in the burned out area. (D) Excavated S. brachyodon plant showing underground stolons.
In addition to S. officinalis and S. fruticosa, in the coastal Adriatic region, a single additional member of the section Salvia can be found: Salvia brachyodon Vandas, or short-toothed sage (Fig. 1). Unlike common sage and Greek sage, S. brachyodon is a narrowly endemic species and can be found in only two localities: St. Ilija, the highest peak of the Pelješac Peninsula (Croatia), and Mt. Orjen, located 120 km to the southeast, in the border area of Bosnia and Hercegovina and Montenegro (Barbalić 1956; Abadžić and Šilić 1982; Šilić 1984). Because of its very limited distribution, habitat fragmentation, obvious ecological succession and other potential environmental threats (e.g. wildfires), short-toothed sage is believed to be a near-threatened species in Croatia (Flora Croatica Database; http://hirc.botanic.hr/fcd) and a highly vulnerable species in Montenegro (Petrović et al. 2008). Unfortunately, to the best of our knowledge, there have been no studies focused on this species for any reason other than its essential oil content (Šavikin-Fodulović et al. 2002; Tzakou et al. 2003), so numerous questions regarding the ecological status and genetic condition of this species' populations have yet to be answered.
To perform comparative population genetic research on selected species growing in an area characterized by intriguing climatological and ecological history and under strong and long-lasting anthropogenic influence (Blondel 2006; Zeder 2008), recently developed SSR markers (Molecular Ecology Resources Primer Development Consortium et al. 2010; Radosavljević et al. 2011, 2012) were applied. The goals of this study were to investigate (i) the consequences of the bottleneck effect and asexual reproduction on population genetic structure, (ii) which population genetic mechanisms could be of help in bypassing inbreeding depression, (iii) how population expansion can be detected on a genetic level and (iv) the possible consequences of inter-species hybridization. We hypothesized that: (1) selected populations of S. officinalis from the geographical centre of the distribution range are characterized by high levels of genetic diversity, are not suffering from any significant population genetic disturbances (e.g. accumulating mutations, genetic drift, inbreeding, population bottleneck etc.) and can consequently serve as a ‘control group’ in contrast to populations of the other two species, S. brachyodon and S. fruticosa), (2) a population of narrow endemic and endangered S. brachyodon is characterized by lower levels of genetic diversity than its widespread congener, i.e. S. officinalis, (3) a presumably non-native and disjunct population of S. fruticosa is genetically severely impoverished and suffers from population bottleneck, inbreeding and the absence of gene flow.
Methods
Leaf tissue from a total of 96 individuals from four populations (24 individuals per population) was collected in two localities: Pelješac Peninsula and the Island of Vis. S. officinalis was collected from both sites, S. brachyodon from the Pelješac Peninsula and S. fruticosa from the Island of Vis. It is important to note that in the studied locations, S. officinalis occurs sympatrically with both taxa and even has an overlapping flowering period with S. fruticosa (late April through early May). The flowering period of S. brachyodon occurs in August and does not overlap with that of S. officinalis (I. Radosavljević, Z. Satovic and Z. Liber, pers. obs.). Voucher specimens of studied populations have been deposited in the Herbarium Croaticum (ZA) of the Department of Botany, Faculty of Science, University of Zagreb.
Genomic DNA was extracted from silica gel-dried leaf tissue by using a GenElute Plant Genomic DNA Miniprep Kit (Sigma-Aldrich, St. Louis, MO, USA). A microsatellite analysis using 13 sequence-tagged SSR loci (Table 1) was performed according to Radosavljević et al. (2012). All microsatellite markers were isolated from S. officinalis and tested for cross-amplification in S. fruticosa and S. brachyodon, but with limited success (Radosavljević et al. 2011, 2012). Due to this difficulty, we were unable to perform the entire analysis using a common set of SSR markers. Instead, we used three sets with the maximum possible number of common markers for each analysed combination of species. SSR amplification reactions were performed in a total volume of 20 μL containing 10× PCR buffer, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.075 μM TAIL FOR primer, 0.2 μM TAIL REV primer, 0.2 μM M13 primer and 0.5 U of Taq HS polymerase (Takara Bio Inc., Shiga, Japan). The amplification was performed with a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) using a two-step PCR protocol with an initial touchdown cycle. The cycling conditions were as follows: 94 °C for 5 min; five cycles of 45 s at 94 °C, 30 s of annealing, beginning at 60 °C and lowered by 1 °C in each cycle, and 90 s at 72 °C; 25 cycles of 45 s at 94 °C, 30 s at 55 °C, and 90 s at 72 °C; and an 8-min extension step at 72 °C. The amplification products were run on an ABI 3730XL analyzer (Applied Biosystems) and the results were analysed with GeneMapper 4.0 software (Applied Biosystems).
Table 1.
Population genetic parameter estimates for each microsatellite locus surveyed in four populations of S. officinalis, S. brachyodon and S. fruticosa. Na, number of alleles; HO, observed heterozygosity; HE, expected heterozygosity; FIS, inbreeding coefficient. Significant deviations from Hardy–Weinberg proportions after sequential Bonferroni corrections: ***significance at the 0.1 % nominal level; **significance at the 1 % nominal level; *significance at the 5 % nominal level. N, number of individuals; G, number of genotypes; Sb, S. brachyodon population; So1, S. officinalis population from the Pelješac Peninsula; So2, S. officinalis population from the Island of Vis and Sf—S. fruticosa population.
| Locus name | Na | HO | HE | FIS | Sign. |
|---|---|---|---|---|---|
| Pelješac Peninsula—S. brachyodon (N = 24, G = 22) | |||||
| SoUZ001 | 4 | 0.750 | 0.691 | −0.085 | ns |
| SoUZ002 | 3 | 0.667 | 0.548 | −0.217 | ns |
| SoUZ005 | 5 | 0.826 | 0.787 | −0.050 | ns |
| SoUZ006 | 9 | 0.792 | 0.861 | 0.080 | ns |
| SoUZ007 | 9 | 0.917 | 0.861 | −0.065 | ns |
| SoUZ008 | 3 | 0.833 | 0.659 | −0.266 | ns |
| SoUZ011 | 6 | 0.917 | 0.788 | −0.163 | ns |
| SoUZ014 | 5 | 0.783 | 0.793 | 0.013 | ns |
| Mean | 5.50 | 0.811 | 0.748 | −0.084 | * |
| Pelješac Peninsula—S. officinalis (N = G = 24) | |||||
| SoUZ001 | 16 | 0.875 | 0.909 | 0.037 | ns |
| SoUZ003 | 9 | 0.833 | 0.842 | 0.010 | ns |
| SoUZ007 | 6 | 0.542 | 0.536 | −0.010 | ns |
| SoUZ009 | 10 | 1.000 | 0.817 | −0.224 | * |
| SoUZ011 | 19 | 0.833 | 0.955 | 0.127 | ns |
| SoUZ013 | 7 | 0.792 | 0.756 | −0.047 | ns |
| SoUZ014 | 15 | 0.958 | 0.913 | −0.050 | ns |
| SoUZ020 | 7 | 0.478 | 0.756 | 0.367 | *** |
| Mean | 11.13 | 0.790 | 0.811 | 0.026 | * |
| Island of Vis—S. officinalis (N = G = 24) | |||||
| SoUZ001 | 10 | 0.875 | 0.828 | −0.057 | ns |
| SoUZ003 | 7 | 0.583 | 0.652 | 0.106 | ns |
| SoUZ007 | 6 | 0.783 | 0.717 | −0.091 | ns |
| SoUZ009 | 4 | 0.833 | 0.667 | −0.250 | ns |
| SoUZ011 | 8 | 0.750 | 0.686 | −0.094 | ns |
| SoUZ013 | 6 | 0.826 | 0.760 | −0.087 | ns |
| SoUZ014 | 8 | 0.870 | 0.840 | −0.035 | ns |
| SoUZ020 | 6 | 0.208 | 0.395 | 0.473 | ** |
| Mean | 6.88 | 0.714 | 0.692 | −0.033 | ns |
| Island of Vis—S. fruticosa (N = G = 24) | |||||
| SoUZ003 | 5 | 0.550 | 0.713 | 0.229 | ns |
| SoUZ005 | 3 | 0.143 | 0.224 | 0.362 | ns |
| SoUZ007 | 3 | 0.238 | 0.219 | −0.087 | ns |
| SoUZ009 | 2 | 0.143 | 0.136 | −0.053 | ns |
| SoUZ013 | 3 | 0.500 | 0.567 | 0.118 | ns |
| SoUZ014 | 2 | 0.191 | 0.176 | −0.081 | ns |
| SoUZ016 | 10 | 0.619 | 0.707 | 0.125 | ns |
| SoUZ020 | 3 | 0.095 | 0.094 | −0.013 | ns |
| Mean | 3.88 | 0.307 | 0.351 | 0.125 | * |
| P (Sb/So1) | 0.010 | 0.635 | 0.293 | ||
| P (Sf/So2) | 0.012 | 0.005 | 0.016 | ||
| P (So1/So2) | 0.056 | 0.370 | 0.074 | ||
GENEPOP 4.0 (Raymond and Rousset 1995) was used to estimate population genetic parameters (the average number of alleles per locus, Na; the observed heterozygosity, HO; the expected heterozygosity or gene diversity, HE; and inbreeding coefficient, FIS) and to test the population genotypic frequencies across all loci for their conformity to Hardy–Weinberg expectations (HWE) (multilocus test). Estimates of Na, HO and HE in each population were compared using the Kruskal–Wallis test in SAS version 8.02 (SAS Institute Inc., Cary, NC, USA). MICRO-CHECKER v.2.2.3 (van Oosterhout et al. 2004) was used to check for scoring errors caused by stutters or large-allele dropouts and to estimate null-allele frequencies. BOTTLENECK program version 1.2.02 (Cornuet and Luikart 1996; Piry et al. 1999) was used to test for evidence of recent bottleneck events on the basis of this theoretical expectation. The expected gene diversity (HE) was compared with the expected gene diversity at mutation-drift equilibrium (HEQ) and calculated from the observed number of alleles under an intermediate two-phase model (TPM) assuming 30 % infinite allele model (IAM) and 70 % stepwise mutation model (SMM) (Di Rienzo et al. 1994). Based on the number of loci in our dataset, the Wilcoxon signed-rank test (Luikart et al. 1998) was chosen for the statistical analysis of heterozygote excess or deficiency, as recommended by Piry et al. (1999). The genetic distances between pairs of S. officinalis, S. brachyodon and S. fruticosa samples were calculated based on eight microsatellite loci by using the proportion of shared-alleles distances, Dpsa (Bowcock et al. 1994) as implemented in MICROSAT (Minch et al. 1996).
To depict reticulate relationships between S. officinalis and S. fruticosa populations, a NeighborNet diagram was produced from a pairwise distance matrix with SplitsTree4 (Huson and Bryant 2006).
To determine the power of the marker set to discriminate among clones, the unbiased probability of identity (PIunbiased; Waits et al. 2001) and the PI for a population composed only by sibs (PIsibs; Evett and Weir 1998; Taberlet and Luikart 1999) was estimated for each locus by using GIMLET software (Valière 2002). Furthermore, to assess the probability that two samples sharing a multilocus genotype originate from distinct sexual reproductive events (i.e. from different zygotes, thus being different genets) we calculated Psex and Psex(f) using the round-robin method of Parks and Werth (1993) as described by Arnaud-Haond et al. (2007) and implemented in GenClone ver. 2.0 (Arnaud-Haond and Belkhir 2007).
To identify putative hybrids between S. fruticosa and S. officinalis, a model-based clustering method was applied on multilocus microsatellite data using the software STRUCTURE ver. 2.3.3 (Pritchard et al. 2000). Ten runs per cluster (K) ranging from 1 to 6 were carried out on the Isabella computer cluster at the University of Zagreb, University Computing Centre (SRCE). Each run consisted of a burn-in period of 200 000 steps followed by 106 MCMC (Monte Carlo Markov Chain) replicates assuming admixture model and correlated allele frequencies. No prior information was used to define the clusters. The most likely number of clusters (K) was chosen by comparing the average estimates of the likelihood of the data, ln[Pr(X|K)], for each value of K (Pritchard et al. 2000), as well as by calculating an ad hoc statistic ΔK, based on the rate of change in the log probability of data between successive K values as described by Evanno et al. (2005). The program STRUCTURE HARVESTER v0.6.92 was used to process the STRUCTURE results files (Earl and vanHoldt 2012).
The Bayesian method implemented by NEWHYBRIDS 1.1. (Anderson and Thompson 2002) was used to assign individuals into six classes: two pure (parental S. fruticosa and S. officinalis) and four hybrids (F1, F2 and backcrosses with the parental populations). The program was run without any prior information about the hybrid status of collected individuals, and with the uniformative Jeffreys prior option for both mixing proportions and allele frequencies. The results were based on the average of five independent runs consisting of a burn-in phase of 100 000 steps, and 500 000 MCMC sweeps. Following the suggestions of Anderson and Thompson (2002), individual genetic assignment to classes was based on a minimum posterior probability threshold (Tq) of 0.50.
Results
The S. officinalis population from the Island of Vis showed no significant deviations from HWE, while the population from the Pelješac Peninsula was characterized by an overall FIS value that was significantly different from zero (FIS = 0.026, P = 0.029). However, a closer inspection revealed that this difference was largely explained by a highly significant FIS value at only one locus (SoUZ020). No significant differences were noted when comparing the overall genetic parameters of these populations (Table 1). For both populations, the Wilcoxon test yielded balanced results under the TPM model, with no significant patterns of either excess or deficiency in the heterozygosity (Table 2). Low differentiation levels between S. officinalis populations were observed (Fig. 2).
Table 2.
Probabilities of heterozygote excess [P(E)] and deficiency [P(D)] according to a Wilcoxon test under the TPM assuming 30 % of IAM and 70 % of SMM.
| Population | Locality | TPM | TPM |
|---|---|---|---|
| P(D) | P(E) | ||
| S. brachyodon | Pelješac | 1.000 | 0.002 |
| S. officinalis | Pelješac | 0.680 | 0.371 |
| S. officinalis | Vis | 0.320 | 0.727 |
| S. fruticosa | Vis | 0.027 | 0.980 |
Figure 2.
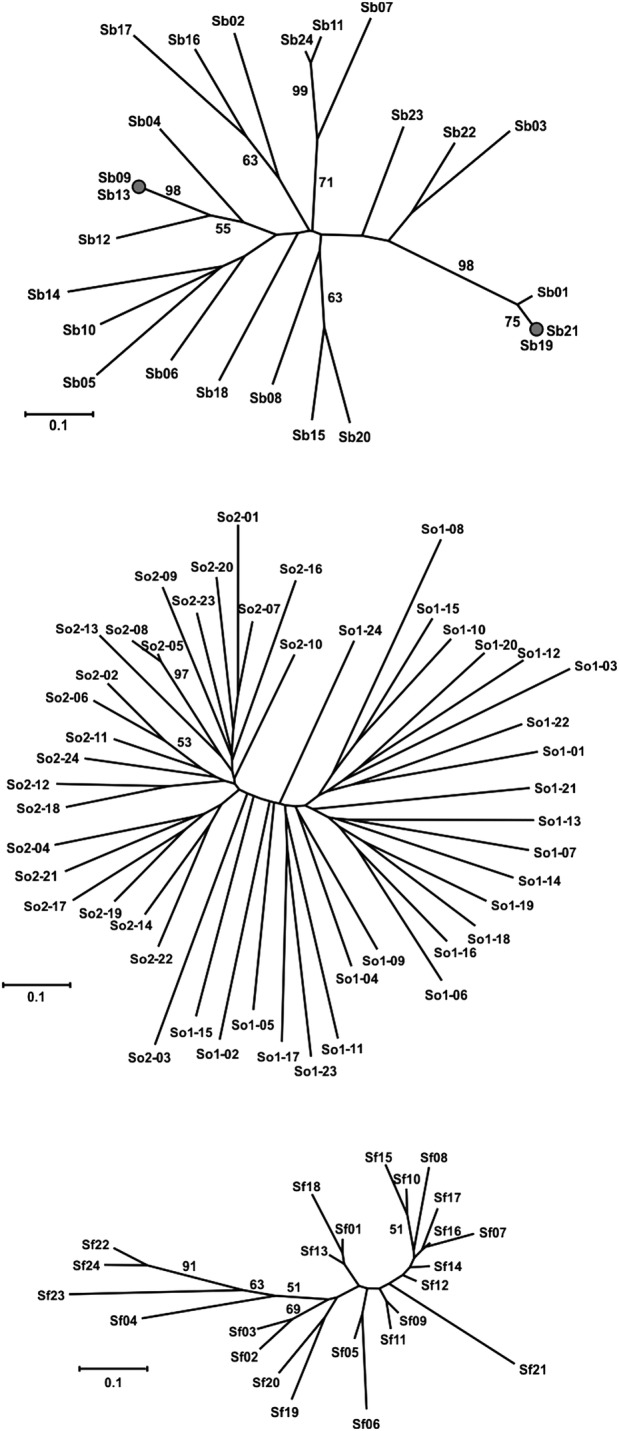
An unrooted Fitch–Margoliash tree based on the proportion-of-shared-allele distances among samples of S. brachyodon, S. officinalis and S. fruticosa (from top to bottom, respectively). Bootstrap support values higher than 50 % for 1000 replicates are indicated on the branches. Salvia officinalis samples marked as ‘So1-’ and ‘So2-’ are from Pelješac Peninsula and the Island of Vis localities, respectively.
In S. brachyodon, the observed heterozygosity was noticeably higher than the expected (0.811 vs. 0.748, respectively), and the FIS value was significantly different from zero (FIS = −0.084, P = 0.013) (Table 1). Under the TPM, the Wilcoxon test revealed significant patterns of excess heterozygosity (P(E) = 0.002) (Table 2). Two pairs of genetically identical individuals were identified from a total of 24 collected individuals (Sb 09/13 and Sb 19/21) (Fig. 2). Eight microsatellite loci used in the study of S. brachyodon population were sufficiently polymorphic to allow individual identification from among 800 × 106 individuals based on the estimate of the unbiased PI (PIunbiased = 1.25 × 10−9) or among 1.286 individuals by assuming that a population was composed only of sibs (PIsibs = 7.77 × 10−4). The probabilities of obtaining the same multilocus genotypes through distinct sexual recombination events were low in both cases (Psex = 5.83 × 10−7 and 4.97 × 10−8, respectively), and even when using a more conservative estimate [Psex(f)] that takes into account possible departures from HWE, the test yielded extremely low values [Psex(f) = 1.04 × 10−6 and 1.25 × 10−7, respectively].
In comparison with the S. officinalis population from the same location, no significant differences between observed and expected heterozygosity were found.
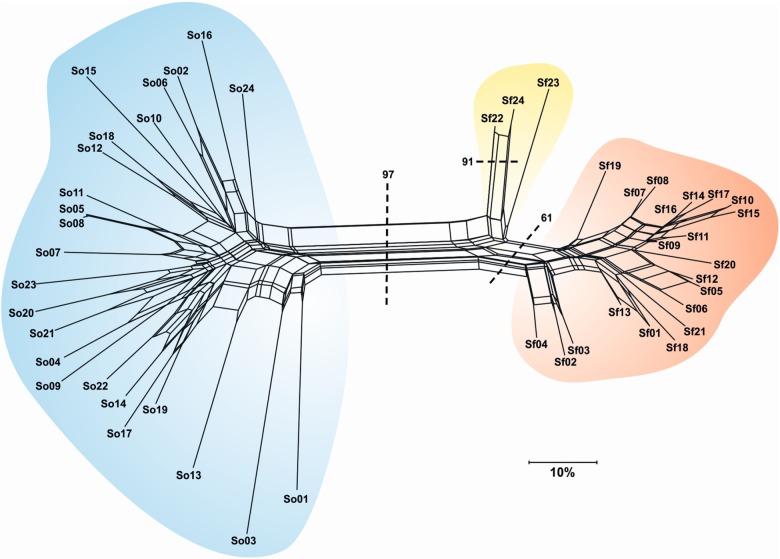
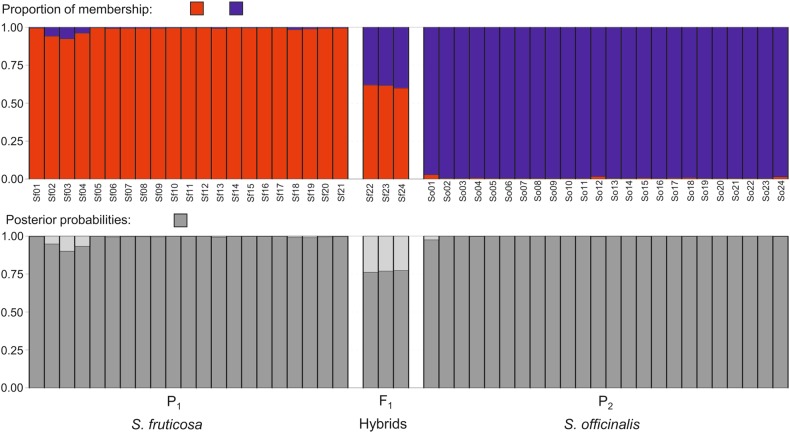
In the S. fruticosa, the observed heterozygosity was lower than expected (0.307 vs. 0.351, respectively), which resulted in a significantly positive FIS value (FIS = 0.125, P= 0.044). Under the TPM, the Wilcoxon test revealed a pattern of heterozygosity deficiency (P(D) = 0.027) (Table 2). When comparing the overall population parameters of S. fruticosa and sympatric S. officinalis, the S. fruticosa presented significantly lower values of the number of alleles, observed and expected heterozygosity (P = 0.012, 0.005 and 0.016, respectively) (Table 1). The close genetic relationship between individuals was confirmed (Fig. 2). In addition to S. officinalis and S. fruticosa, a third group of intermediate individuals was detected, suggesting hybridization between these species (Figs 3 and 4). The results from the Bayesian analysis implemented in STRUCTURE confirmed the existence of two separate clusters (i.e. parental species) and a group of three individuals characterized by admixed proportions (Fig. 4). The highest ΔK was obtained at K = 2 (1862.93), while the second highest ΔK was at K = 4 (9.62). The presence of two well-differentiated populations belonging to parental taxa was strongly supported by the above results, as was the existence of individuals of hybrid origin.
Figure 3.
A neighborNet diagram based on the proportion-of-shared-alleles distance matrix among individuals belonging to S. officinalis (in blue), S. fruticosa (in orange) and their hybrid, S. × auriculata (in yellow). The bootstrap support value was derived from a Neighbor Joining analysis.
Figure 4.
Proportions of membership of each individual in each of the two clusters as estimated by the program STRUCTURE. Each individual plant is represented by a single vertical line divided into colours. Each colour represents one cluster, and the length of the coloured segment shows the individual's estimated proportion of membership in that cluster. Assignment of individuals into classes (parental S. officinalis and S. fruticosa and F1) based on maximum posterior probabilities that each individual belongs to a particular class as estimated by the program NEWHYBRIDS.
The hybrids assignment by NEWHYBRIDS was in concordance with the results from the STRUCTURE (Fig. 4). All individuals characterized by admixed proportions were confirmed as F1 hybrids with very similar posterior probabilities ranging from 0.762 to 0.773. Interestingly, posterior probabilities suggesting the possible origin of these individuals through backcrossing were relatively high and ranged from 0.186 to 0.212.
Discussion
S. officinalis
In the S. officinalis from Pelješac, most loci were in HWE, with the exception of locus SoUZ011, which was characterized by a significantly positive FIS value (Table 1). These results are consistent with the expectation that this population, located in the central part of the eastern Adriatic mainland coastal region where common sage grows abundantly, is in mutation-drift equilibrium and is not experiencing any population genetic disturbances. The results for the population from the Island of Vis were very similar; deviations from HWE were insignificant, and there were no statistically significant population genetic differences between these two populations (Table 1). Being located on an offshore island did not cause a substantial disturbance in the genetic structure of this population. Plausible explanations for this situation may lie in the fact that some 20 000 years ago, during the Last Glacial Maximum, when sea level was ∼120 m lower than today (Rohling et al. 1998; Clark et al. 2009), the Island of Vis was part of the mainland. After the sea level rose, the remaining population was simply large enough to retain most of the genetic variability that has been preserved until the present day. The low level of differentiation between the two S. officinalis populations (Fig. 2) is not surprising because the Pelješac Peninsula is the nearest mainland to the Island of Vis.
Being a typical heliophyte and one of the most abundant plant species in open habitats along the eastern Adriatic coast, S. officinalis is not threatened currently. The selected populations located in the geographical centre of the species distribution area are characterized by high levels of genetic variability and are not experiencing any population genetic disruptive circumstances. Because the results confirm hypothesis (1), S. officinalis populations can be treated as reference populations in contrast to populations of the other two species that probably experienced some of these disruptive factors.
S. brachyodon
In comparison to the S. officinalis from the same location, S. brachyodon was characterized by no significant differences between the observed and expected levels of heterozygosity. There are several possible explanations for such high heterozygosity levels and heterozygosity excess (negative FIS value). First, during the sampling expedition, a large number of bumblebees (Bombus sp.) were observed to visit S. brachyodon flowers, supporting the expectation that this species, like most sage species with large flowers (Haque and Ghoshal 1981), is predominantly an outbreeding species. Second, although the large and branched inflorescences that usually grow over 1 m in height produce a significant number of flowers, they tend to bloom sequentially (basitonic type), resulting in very few flowers on the same plant being available for pollination simultaneously (I. Radosavljević, Z. Satovic and Z. Liber, pers. obs.), thus favouring pollination between different individuals. These two traits may help to maintain genetic variation by preventing self-pollination; that is, by promoting outcrossing, the probability of creating homozygous offspring is reduced. Third, S. brachyodon tends not to grow individually but rather in dense groups of individuals, which indicates clonal reproduction by stolons (Fig. 1). Within our sample set of 24 individuals, two sample pairs were shown to be genetically identical (Fig. 2), thus supporting our observation. Additional conformation that clonality, rather than high inbreeding levels, is responsible for the presence of individuals sharing the same multilocus genotype, was obtained by GenClone software. With clonality being confirmed, the negative FIS value, e.g. heterozygosity excess, could easily be explained (Delmotte et al. 2002; Balloux et al. 2003; Alberto et al. 2005; Ruggiero et al. 2005). As stated several times (Judson and Normark 1996; Welch and Meselson 2000), asexual reproduction can be responsible for maintaining high heterozygosity levels or even for increasing heterozygosity by the accumulation of mutations over generations. New mutations that occur are permanently fixed and cannot be lost through genetic drift because of the presence of non-sexual reproduction.
The BOTTLENECK test provided evidence that this S. brachyodon population has recently experienced severe reductions in the effective population size, i.e. a bottleneck. There are two possible explanations for a population bottleneck that should be taken into account: wildfire and ecological succession. Twelve years before sampling, in August of 1998, a devastating fire overran the entire area where this population is located (Fig. 1). However, as documented on numerous occasions, through the re-establishment of open, non-shaded habitats, periodic fires in Mediterranean-type ecosystems play a vital role in their evolutionary history (Naveh 1975; Keeley 1977; Ne'eman and Dafni 1999), rather than having a destructive impact on local biodiversity. Nevertheless, human-induced fires are prevalent in contrast to naturally occurring fires, as only 1–5 % of all the wildfires in the Mediterranean are of natural origin (Alexandrian et al. 1998). Thus, it is very likely that this specific fire was also human-induced. However, the presence of this isolated population suggests that S. brachyodon, being a native Mediterranean species, is well adapted to such an extreme event and it is unlikely that the fire caused any significant contraction in population size. Although the species cannot be characterized as a hemicryptophyte but rather as a chamaephyte, due to its clonal reproduction and the presence of underground stolons, it is also likely that resprouting is a possible mechanism of post-fire regeneration (Naveh 1975; Keeley 1977; Lloret et al. 2003). It is also possible that S. brachyodon is, such as S. fruticosa, an obligate seeder (Ne'eman and Dafni 1999), and may be dependent on a soil-stored seed bank. For such species, in the absence of human activities that support open-type habitats (e.g. certain styles of agriculture), occasional fires are needed to ‘clear’ the terrain of vegetation, consequently allowing seedlings to establish (Ne'eman and Dafni 1999; Santana et al. 2012). Once pastures, today the karstic plains in the Pelješac Peninsula are mostly abandoned; consequently, gradual ecological succession towards the indigenous black pine (Pinus nigra Arnold) forest (Flora Croatica Database; http://hirc.botanic.hr/fcd) is common and inevitable throughout the region. Being a typical heliophyte, S. brachyodon requires clear, open space to thrive and the abandonment of the traditional pasture management may result in a loss of a suitable habitats. The ecological succession at this location prior to the 1998 fire is the most likely reason for this population bottleneck and recent wildfire likely saved this population from deterioration and possibly even extinction. To re-establish and maintain the desired open habitat and the associated species community that emerged from the wildfire, grazing cattle could be reintroduced in such areas (Verdú et al. 2000).
Although narrow endemic species in general are considered to be characterized by low levels of genetic diversity (e.g. Gitzendanner and Soltis 2000), on some occasions it has proven to be the opposite (e.g. Dolan et al. 1999; Martinez-Palacios et al. 1999; Hannan and Orick 2000). Therefore, the high levels of genetic variation found in S. brachyodon that failed to support the predicted hypothesis (2) are not unprecedented. Clonal reproduction can serve as a valuable mechanism to preserve genetic diversity and in spite of the detected population bottleneck, high levels of genetic variability can indeed be found in this species.
S. fruticosa
Two parameters that are interesting for this isolated population are the positive and highly significant FIS value, which suggests high inbreeding levels, and the significant heterozygosity deficiency as revealed by BOTTLENECK. The results additionally confirm the hypothesis of the non-native origin of S. fruticosa in this location. As is often the case with anthropogenic introductions, this population was most likely founded from a very limited number of individuals who were completely isolated from the source population and from any other population of their kind. The resulting genetic drift, bottleneck and inevitable breeding between close relatives subsequently led to increased homozygosity, which had a significantly higher value when compared with the sympatric S. officinalis population (Table 1). Furthermore, if our hypothesis was correct and assuming that this introduction was intentional due to the settlers' (i.e. Ancient Greeks) knowledge of the aromatic and health-beneficial properties of this species (Rivera et al. 1994), it is reasonable to assume that this population was anthropogenically restrained from expansion and was composed of a very limited number of individuals for a long period of time. During this prolonged bottleneck period, S. fruticosa was grown only in gardens and vineyards surrounding local settlements. The expansion of S. fruticosa took place just recently, in approximately the last hundred years, when much of the local population emigrated, resulting in the abandonment of traditional agriculture. This hypothesis is indeed consistent with the current distribution of S. fruticosa on the Island of Vis because it is found exclusively in open, non-shaded habitats of neglected and abandoned vineyards and olive groves surrounding local settlements. The fact that S. fruticosa is a native East-Mediterranean species has eased its adaptation to a new environment that did not differ significantly from that in its native area of distribution, allowing S. fruticosa to expand quickly and form a current population of a few thousand individuals. In addition, the close genetic relationship within the population that can be observed today (Fig. 2) supports the hypothesis that this population was started relatively recently by a very limited number of individuals. During this relatively short period, there was not enough time, i.e. not enough generations, for newly emerging mutations to spread through the population and to accumulate in higher frequencies. Thus, we are now observing a significant heterozygosity deficiency (HE< HEQ). It is likely that over time and within a population of this size, the frequency of new alleles, and consequently heterozygosity, will increase.
However, considering all the results obtained in this study, it is becoming clearer that favourable ecological parameters are not the only factors responsible for such a vigorous and successful expansion of S. fruticosa. As already mentioned, S. officinalis and S. fruticosa occur sympatrically in this location with overlapping flowering periods. It is also important to note that a putative hybrid between these species has been recorded (Salvia × auriculata Mill.) (Putievsky et al. 1990), but to the best of our knowledge, only as a result of artificial crossing (Putievsky et al. 1990; Dudai et al. 1999; Reales et al. 2004). It is also noteworthy that three sampled specimens of S. fruticosa were characterized by non-typical morphological traits for this species, especially the calyx shape (non-radial, slightly zygomorphic) and the calyx trichome length and density (which were more pronounced than usual). However, because this species is characterized by exceptionally high levels of morphological variability (Hedge 1972; Karousou and Kokkini 1997), these samples were considered to originate from S. fruticosa. As revealed by the computer programs SplitsTree (Fig. 3), STRUCTURE and NEWHYBRIDS (Fig. 4) all three doubtful individuals (Sf22, Sf23 and Sf24) were of hybrid origin. Although the results undoubtedly support the origin of these individuals through hybridization, their asymmetric positioning in the neighbour-net diagram (Fig. 3) and somewhat inconclusive results from NEWHYBRIDS (Fig. 4) also suggest their possible origin through backcrossing with one of the parental species, i.e. S. fruticosa. However, the sample set is not large enough for more detailed and comprehensive study of the hybridization event, so it is impossible to address some very important questions, such as (i) are the hybrids fertile, (ii) is backcrossing to either parental species indeed present and (iii) if backcrossing is present, is it symmetrical or unilateral etc. As hybridization between species is generally considered to be rare on an individual basis, a very limited number of individuals participate in hybridization events. Consequently, hybrids are rare in the population, but even a few hybrids can provide a bridge that allows a trickle of alleles to pass between species (Mallet 2005). Thus, if we leave the possibility of the backcrossing with S. fruticosa open, the appearance of certain low-frequency alleles in S. fruticosa is not surprising. As new alleles originating from S. officinalis reach S. fruticosa through backcrossing, the alleles can be found in only a small percentage of individuals. Consequently, the frequency of these genes in the new gene pool is very low. They therefore contribute considerably to increased allelic diversity but not to heterozygosity, which is consequently detected by BOTTLENECK as heterozygosity deficiency (Table 2). However, as S. officinalis and S. fruticosa are undoubtedly closely related species it is possible to assume that at least some of these rare common alleles are not necessarily the result of inter-species introgression and horizontal gene transfer, but instead share the same origin through a common ancestor. It is reasonable to assume that the high adaptability as well as the genetic and range expansion that can be observed in the S. fruticosa population is supported by introgression and hybridization with a successful and well-adapted member of the indigenous flora, S. officinalis.
In addition, naturally occurring hybridization between these species may be of significant agricultural interest. As mentioned above, artificial crossing between these species has been already performed, and comparative research should address the yield and content of essential oil from hybrid individuals of artificial and natural origin.
The results support the hypothesis (3) that the S. fruticosa population is characterized by very low levels of genetic variability. As a bottleneck event can be detected in a limited time frame after it happens (Peery et al. 2012), we were unable to recognize it. However, high levels of inbreeding accompanied by very low levels of genetic variability suggested the occurrence of a bottleneck event in the past. The inter-population gene flow, expected to be absent in remote and isolated populations, was possibly present, but on an inter-species level. Hybridization with indigenous S. officinalis was confirmed, but its nature and direction are still uncertain.
Conclusions
Our initial prediction regarding the levels of genetic diversity of three sympatrically growing Salvia species were only partially confirmed. In accordance with expectations, S. officinalis populations, as true core populations, were characterized by high levels of variability, while the disjunct and isolated population of S. fruticosa contained significantly less genetic variation and struggled with high levels of inbreeding. The natural hybridization between S. officinalis and S. fruticosa was confirmed for the first time. To assess the nature and direction of hybridization, more extensive research including a significantly larger number of samples and additional methods should be conducted. Although morphometrical analyses alone are considered to be of limited value in hybrid identification and characterization (Rieseberg et al. 1993), if combined with molecular methods, e.g. SSRs, more informative and insightful results could be obtained.
In contrast to expectation, the population of the endangered and narrow endemic S. brachyodon, even after surviving a population bottleneck, was still characterized by surprisingly high levels of genetic diversity. Detected clonal reproduction via underground stolons could serve as a valuable mechanism and adaptation in preservation and even accumulation of emerging mutations, although a larger sample set and appropriate analysis methods are needed for more comprehensive results and conclusions. Detailed spatial-genetic analysis would give an inside perspective on this crucial life trait (i.e. clonality) as numerous questions dealing with this species' historical and contemporary demography could be answered.
Some of the detected fluctuations in genetic structures of the studied Salvia populations could also be a result of certain human activities or their absence. However, before reaching any conclusions, caution is needed, as some detected genetic traits could easily be a result of specific differences in species life history traits, rather than a consequence of human activities. To more reliably address this and similar topics, a different approach is needed.
This research demonstrates the importance and advantages of a multi-species approach in population genetics studies, as some relevant findings and conclusions about each species can be reached only if a comparison among different, yet similar and closely related species is performed. The selection of the ‘control’ group should be carefully considered, as it will set the frame of the entire research. Presumably, widespread species with no significant population genetic disturbances, highly specific life traits or large-scale demographic fluctuations, should be considered as the right choice for such a purpose.
Sources of Funding
This study was supported by the Croatian Science Foundation within the framework of the Project No. 09.01/246 (Epigenetic vs genetic diversity in natural plant populations: A case study of Croatian endemic Salvia species).
Contributions by the Authors
I.R. performed the experiment, wrote and edited the manuscript and created all the figures. Z.S. conducted statistical analyses. Z.L. conceived the idea for the research and extensively edited the manuscript. All authors were equally involved in sample collection.
Conflict of Interest Statement
None declared.
Acknowledgements
The authors thank two anonymous reviewers and an Associate Editor for their useful comments that substantially improved this manuscript. Our sincere gratitude is also expressed to our colleague, Boštjan Surina, for his comments on the previous versions of the manuscript.
Literature Cited
- Abadžić S, Šilić Č. 1982. Chorology, ecology and phytosociological affiliation of Salvia brachyodon Vandas in flora of Yugoslavia (in Montenegrin). Glasnik Republičkog Zavoda za Zaštitu Prirode i Prirodnjačkog Muzeja u Titogradu 15:125–131. [Google Scholar]
- Alberto F, Gouveia L, Arnaud-Haond S, Pérez-Lloréns JL, Duarte CM, Serrão EA. 2005. Within-population spatial genetic structure, neighbourhood size and clonal subrange in the seagrass Cymodocea nodosa. Molecular Ecology 14:2669–2681. 10.1111/j.1365-294X.2005.02640.x [DOI] [PubMed] [Google Scholar]
- Alexandrian D, Esnault F, Calabri G. 1998. Forest fires in the Mediterranean area. Food and Agriculture Organization of the United Nations, The Forestry Department. [Google Scholar]
- Anderson EC, Thompson EA. 2002. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud-Haond S, Belkhir K. 2007. GENCLONE 1.0: a new program to analyze genetics data on clonal organism. Molecular Ecology Notes 7:15–17. [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA. 2007. Standardizing methods to address clonality in population studies. Molecular Ecology 16:5115–5139. 10.1111/j.1365-294X.2007.03535.x [DOI] [PubMed] [Google Scholar]
- Arnold ML. 1997. Natural hybridization and evolution. Oxford: Oxford University Press. [Google Scholar]
- Balloux F, Lehmann L, de Meeus T. 2003. The population genetics of clonal and partially clonal diploids. Genetics 164:1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalić Lj. 1956. Contribution to the knowledge of Salvia brachyodon Vand (in Croatian). Periodicum Biologorum 9:5–10. [Google Scholar]
- Blondel J. 2006. The ‘design’ of Mediterranean landscapes: a millennial story of humans and ecological systems during the historic period. Human Ecology 34:713–729. 10.1007/s10745-006-9030-4 [DOI] [Google Scholar]
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. 1994. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457. 10.1038/368455a0 [DOI] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM. 2009. The last glacial maximum. Science 325:710–714. 10.1126/science.1172873 [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G. 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutini M, Cancellieri L, Ceschin S, Lucchese F, Caneva G. 2007. Analisi cenologica e sintassonomia delle garighe a Salvia officinalis L. lucane nel quadro dei salvieti peninsulari (Basilicata, Appennino meridionale). Webbia 62:225–244. [Google Scholar]
- Delmotte F, Leterme N, Gauthier J-P, Rispe C, Simon J-C. 2002. Genetic architecture of sexual and asexual populations of the aphid Rhopalosiphum padi based on allozyme and microsatellite markers. Molecular Ecology 11:711–723. 10.1046/j.1365-294X.2002.01478.x [DOI] [PubMed] [Google Scholar]
- Di Pietro R. 2011. New dry grassland associations from the Ausoni-Aurunci mountains (central Italy)—syntaxonomical updating and discussion on the higher rank syntaxa. Hacquetia 10:183–231. 10.2478/v10028-011-0011-9 [DOI] [Google Scholar]
- Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB. 1994. Mutational processes of simple-sequence repeat loci in human populations. Proceedings of the National Academy of Sciences of the USA 91:3166–3170. 10.1073/pnas.91.8.3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan RW, Yahr R, Menges ES, Halfhill MD. 1999. Conservation implications of genetic variation in three rare species endemic to Florida rosemary scrub. American Journal of Botany 86:1556–1562. 10.2307/2656793 [DOI] [PubMed] [Google Scholar]
- Dudai N, Lewinsohn E, Larkov O, Katzir I, Ravid U, Chimovitsh D, Sa'ad D, Putievsky E. 1999. Dynamics of yield components and essential oil production in a commercial hybrid sage (Salvia officinalis × Salvia fruticosa cv. Newe Ya'ar No. 4). Journal of Agricultural and Food Chemistry 47:4341–4345. 10.1021/jf9901587 [DOI] [PubMed] [Google Scholar]
- Earl DA, vanHoldt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. 2008. Genetic variation across species’ geographical ranges: the central–marginal hypothesis and beyond. Molecular Ecology 17:1170–1188. 10.1111/j.1365-294X.2007.03659.x [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14:2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Evett I, Weir B. 1998. Interpreting DNA evidence: statistical genetics for forensic scientists. Sunderland: Sinauer Associates. [Google Scholar]
- Flora Croatica Database. http://hirc.botanic.hr/fcd (21 May 2013).
- Fonderflick J, Lepart J, Caplat P, Debussche M, Marty P. 2010. Managing agricultural change for biodiversity conservation in a Mediterranean upland. Biological Conservation 143:737–746. 10.1016/j.biocon.2009.12.014 [DOI] [Google Scholar]
- Gaston KJ. 2003. The structure and dynamics of geographic ranges. Oxford: Oxford University Press. [Google Scholar]
- Gitzendanner MA, Soltis PS. 2000. Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany 87:783–792. 10.2307/2656886 [DOI] [PubMed] [Google Scholar]
- Hannan GL, Orick MW. 2000. Isozyme diversity in Iris cristata and the threatened glacial endemic I. lacustris (Iridaceae). American Journal of Botany 87:293–301. 10.2307/2656625 [DOI] [PubMed] [Google Scholar]
- Haque MS, Ghoshal KK. 1981. Floral biology and breeding system in the genus Salvia L. Proceedings of the National Academy of Sciences, India 47:716–724. [Google Scholar]
- Hedge IC. 1972. Salvia. In: Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA, eds. Flora Europaea, Vol. 3 Cambridge: Cambridge University Press, 188–192. [Google Scholar]
- Hedge IC. 1982. Salvia L. In: Davis PH, ed. Flora of Turkey and the East Aegean Islands, Vol. 7 Edinburgh: Edinburgh University Press, 400–461. [Google Scholar]
- Hewitt GM. 1999. Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society 68:87–112. 10.1111/j.1095-8312.1999.tb01160.x [DOI] [Google Scholar]
- Hewitt GM. 2001. Speciation, hybrid zones and phylogeography—or seeing genes in space and time. Molecular Ecology 10:537–549. 10.1046/j.1365-294x.2001.01202.x [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23:254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Judson OP, Normark BB. 1996. Ancient asexual scandals. Trends in Ecology and Evolution 11:41–46. 10.1016/0169-5347(96)81040-8 [DOI] [PubMed] [Google Scholar]
- Karousou R, Kokkini S. 1997. Distribution and clinal variation of Salvia fruticosa Mill. (Labiatae) on the island of Crete (Greece). Willdenowia 27:113–120. 10.3372/wi.27.2710 [DOI] [Google Scholar]
- Karousou R, Hanlidou E, Kokkini S. 2000. The Sage plants of Greece: distribution and infraspecific variation. In: Kintzios SE, ed. SAGE The genus Salvia. Amsteldijk: Harwood Academic Publishers imprint, part of the Gordon and Breach Publishing Group, 27–46. [Google Scholar]
- Keeley JE. 1977. Seed production, seed populations in soil, and seedling production after fire for two congeneric pairs of sprouting and nonsprouting chaparal shrubs. Ecology 58:820–829. 10.2307/1936217 [DOI] [Google Scholar]
- Lesica P, Allendorf FW. 1995. When are peripheral populations valuable for conservation? Conservation Biology 9:753–760. 10.1046/j.1523-1739.1995.09040753.x [DOI] [Google Scholar]
- Lloret F, Pausas JG, Vilà M. 2003. Responses of Mediterranean Plant Species to different fire frequencies in Garraf Natural Park (Catalonia, Spain): field observations and modelling predictions. Plant Ecology 167:223–235. 10.1023/A:1023911031155 [DOI] [Google Scholar]
- Lucchese F, Persia G, Pignatti S. 1995. I prati a Bromus erectus Hudson dell'Appennino Laziale. Fitosociologia 30:145–180. [Google Scholar]
- Luikart G, Allendorf FW, Cornuet J-M, Sherwin WB. 1998. Distortion of allele frequency distributions provides a test for recent population bottlenecks. Journal of Heredity 89:238–247. 10.1093/jhered/89.3.238 [DOI] [PubMed] [Google Scholar]
- Mallet J. 2005. Hybridization as an invasion of the genome. Trends in Ecology and Evolution 20:229–237. 10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Martinez-Palacios A, Eguiarte LE, Furnier GR. 1999. Genetic diversity of the endangered endemic Agave victoriae-reginae (Agavaceae) in the Chihuahuan desert. American Journal of Botany 86:1093–1098. 10.2307/2656971 [DOI] [PubMed] [Google Scholar]
- Meeus S, Honnay O, Jacquemyn H. 2012. Strong differences in genetic structure across disjunct, edge, and core populations of the distylous forest herb Pulmonaria officinalis (Boraginaceae). American Journal of Botany 99:1809–1818. 10.3732/ajb.1200223 [DOI] [PubMed] [Google Scholar]
- Michaux JR, Magnanou E, Paradis E, Nieberding C, Libois R. 2003. Mitochondrial phylogeography of the woodmouse (Apodemus sylvaticus) in the western Palearctic region. Molecular Ecology 12:685–697. 10.1046/j.1365-294X.2003.01752.x [DOI] [PubMed] [Google Scholar]
- Miller NF. 1992. The origins of plant cultivation in the Near East. In: Cowan CW, Watson PJ, eds. The origins of agriculture: an international perspective. Washington: Smithsonian Institution Press, 39–58. [Google Scholar]
- Minch E, Ruiz-Linares A, Goldstein D, Feldman M, Cavalli-Sforza LL. 1996. Microsat: a computer program for calculating various statistics on microsatellite allele data. Version 1.5 Stanford: Stanford University Medical Center. [Google Scholar]
- Molecular Ecology Resources Primer Development Consortium, An J, Bechet A, Berggren A, Brown SK, Bruford MW, Cai Q, Cassel-Lundhagen A, Cezilly F, Chen S-L, Cheng W, Choi S-K, Ding XY, Fan Y, Feldheim KA, Feng ZY, Friesen VL, Gaillard M, Galaraza JA, Gallo L, Ganeshaiah KN, Geraci J, Gibbons JG., Grant WS., Grauvogel Z, Gustafsson S, Guyon JR., Han L, Heath DD, Hemmilä S, Hogan JD, Hou BW, Jakse J, Javornik B, Kaňuch P, Kim K-K, Kim K-S, Kim S-G, Kim S-I, Kim W-J, Kim Y-K, Klich MA, Kreiser BR, Kwan Y-S, Lam AW, Lasater K, Lascoux M, Lee H, Lee Y-S, Li DL, Li S-J, Li WY, Liao X, Liber Z, Lin L, Liu S, Luo X-H, Ma YH, Ma Y, Marchelli P, Min M-S, Moccia MD, Mohana KP, Moore M, Morris-Pocock JA, Park H-C, Pfunder M, Ivan R, Ravikanth G, Roderick GK., Rokas A, Sacks BN, Saski CA, Satovic Z, Schoville SD, Sebastiani F, Sha Z-X, Shin E-H, Soliani C, Sreejayan N, Sun Z, Tao Y, Taylor SA, Templin WD, Shaanker RU, Vasudeva R, Vendramin GG, Walter RP, Wang G-Z, Wang K-J, Wang YQ, Wattier RA, Wei F, Widmer A, Woltmann S, Won Y-J, Wu J, Xie ML, Xu G, Xu X-J, Ye H-H, Zhan X, Zhang F, Zhong J. 2010. Permanent genetic resources added to molecular ecology resources database 1 October 2009–30 November 2009. Molecular Ecology Resources 10:404–408. 10.1111/j.1755-0998.2009.02827.x [DOI] [PubMed] [Google Scholar]
- Naveh Z. 1975. The evolutionary significance of fire in the Mediterranean region. Vegetatio 29:199–208. 10.1007/BF02390011 [DOI] [Google Scholar]
- Naveh Z, Dan J. 1973. The human degradation of Mediterranean landscapes in Israel. In: di Castri F, Mooney HA, eds. Mediterranean type ecosystems: origin and structure. Berlin: Springer, 372–390. [Google Scholar]
- Ne'eman G, Dafni A. 1999. Fire, bees, and seed production in a Mediterranean key species Salvia fruticosa Miller (Lamiaceae). Israel Journal of Plant Sciences 47:157–163. 10.1080/07929978.1999.10676768 [DOI] [Google Scholar]
- Parks JC, Werth CR. 1993. A study of spatial features of clones in a population of Bracken fern, Pteridium aquilinum (Dennstaedtiaceae). American Journal of Botany 80:537–544. 10.2307/2445369 [DOI] [PubMed] [Google Scholar]
- Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Robinson S, Vásquez-Carrillo C, Pauli JN, Palsbøll PJ. 2012. Reliability of genetic bottleneck tests for detecting recent population declines. Molecular Ecology 21:3403–3418. 10.1111/j.1365-294X.2012.05635.x [DOI] [PubMed] [Google Scholar]
- Petrović D, Stešević D, Vuksanović S. 2008. Materials for the Red book of Montenegro. Natura Montenegrina 7:605–631. [Google Scholar]
- Pignatti S. 1982. Flora d’ Italia, Vol. 2 Bologna: Edagricole. [Google Scholar]
- Piry S, Luikart G, Cornuet JM. 1999. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity 90:502–503. 10.1093/jhered/90.4.502 [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putievsky E, Ravid U, Diwan-Rinzler N, Zohary D. 1990. Genetic affinities and essential oil composition of Salvia officinalis L., S. fruticosa Mill., S. tomentosa Mill. and their hybrids. Flavour and Fragrance Journal 5:121–123. 10.1002/ffj.2730050213 [DOI] [Google Scholar]
- Radosavljević I, Jakse J, Javornik B, Satovic Z, Liber Z. 2011. New microsatellite markers for Salvia officinalis (Lamiaceae) and cross-amplification in closely related species. American Journal of Botany 98:e316–e318. 10.3732/ajb.1000462 [DOI] [PubMed] [Google Scholar]
- Radosavljević I, Satovic Z, Jakse J, Javornik B, Greguraš D, Jug-Dujaković M, Liber Z. 2012. Development of new microsatellite markers for Salvia officinalis L. and its potential use in conservation-genetic studies of narrow endemic Salvia brachyodon Vandas. International Journal of Molecular Sciences 13:12082–12093. 10.3390/ijms130912082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. 1995. GenePop (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86:248–249. [Google Scholar]
- Reales A, Riviera D, Palazon JA, Obon C. 2004. Numerical taxonomy study of Salvia sect. Salvia (Labiatae). Botanical Journal of the Linnean Society 145:353–371. 10.1111/j.1095-8339.2004.00295.x [DOI] [Google Scholar]
- Rieseberg LH, Ellstrand NC, Arnold M. 1993. What can molecular and morphological markers tell us about plant hybridization? Critical Reviews in Plant Sciences 12:213–241. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216. 10.1126/science.1086949 [DOI] [PubMed] [Google Scholar]
- Rivera D, Obon C, Cano F. 1994. The botany, history and traditional uses of three-lobed sage (Salvia fruticosa Miller) (Labiatae). Economic Botany 48:190–195. 10.1007/BF02908216 [DOI] [Google Scholar]
- Rohling EJ, Fenton M, Jorissen FJ, Bertrand P, Ganssen G, Caulet JP. 1998. Magnitudes of sea-level lowstands of the past 500,000 years. Nature 394:162–165. 10.1038/28134 [DOI] [Google Scholar]
- Ruggiero MV, Reusch TBH, Procaccini G. 2005. Local genetic structure in a clonal dioecious angiosperm. Molecular Ecology 14:957–967. 10.1111/j.1365-294X.2005.02477.x [DOI] [PubMed] [Google Scholar]
- Sagarin RD, Gaines SD, Gaylord B. 2006. Moving beyond assumptions to understand abundance distributions across the ranges of species. Trends in Ecology and Evolution 21:524–530. 10.1016/j.tree.2006.06.008 [DOI] [PubMed] [Google Scholar]
- Samis KE, Eckert CG. 2007. Testing the abundant center model using range-wide demographic surveys of two coastal dune plants. Ecology 88:1747–1758. 10.1890/06-1153.1 [DOI] [PubMed] [Google Scholar]
- Santana VM, Baeza MJ, Maestre FT. 2012. Seedling establishment along post-fire succession in Mediterranean shrublands dominated by obligate seeders. Acta Oecologica 39:51–60. 10.1016/j.actao.2011.12.001 [DOI] [Google Scholar]
- Šavikin-Fodulović KP, Tasić SR, Menković NR. 2002. The essential oil of Salvia brachyodon Vandas. Lamiaceae. Journal of Essential Oil Research 14:342–343. 10.1080/10412905.2002.9699876 [DOI] [Google Scholar]
- Seehausen O. 2004. Hybridization and adaptive radiation. Trends in Ecology and Evolution 19:198–207. 10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Šilić Č. 1984. Endemic plants (in Bosnian) Sarajevo: Svjetlost. [Google Scholar]
- Stebbins GL. 1959. The role of hybridization in evolution. Proceedings of the American Philosophical Society 103:231–251. [Google Scholar]
- Surina B, Schneeweiss GM, Glasnović P, Schönswetter P. 2014. Testing the efficiency of nested barriers to dispersal in the Mediterranean high mountain plant Edraianthus graminifolius (Campanulaceae). Molecular Ecology 23:2861–2875. 10.1111/mec.12779 [DOI] [PubMed] [Google Scholar]
- Taberlet P, Luikart G. 1999. Non-invasive genetic sampling and individual identification. Biological Journal of the Linnean Society 68:41–55. 10.1111/j.1095-8312.1999.tb01157.x [DOI] [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F. 1998. Comparative phylogeography and postglacial colonization routes in Europe. Molecular Ecology 7:453–464. 10.1046/j.1365-294x.1998.00289.x [DOI] [PubMed] [Google Scholar]
- Tzakou O, Couladis M, Slavkovska V, Mimica-Dukic N, Jancic R. 2003. The essential oil composition of Salvia brachyodon Vandas. Flavour and Fragrance Journal 18:2–4. 10.1002/ffj.1132 [DOI] [Google Scholar]
- Valière N. 2002. GIMLET: a computer program for analysing genetic individual identification data. Molecular Ecology Notes 2:377–379. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. 2004. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4:535–538. 10.1111/j.1471-8286.2004.00684.x [DOI] [Google Scholar]
- Vasić O. 1997. A survey of the Mediterranean species of Lamiaceae family in the flora of Serbia. Lagascalia 19:263–270. [Google Scholar]
- Verdú JR, Crespo MB, Galante E. 2000. Conservation strategy of a nature reserve in Mediterranean ecosystems: the effects of protection from grazing on biodiversity. Biodiversity and Conservation 9:1707–1721. 10.1023/A:1026506725251 [DOI] [Google Scholar]
- Waits LP, Luikart G, Taberlet P. 2001. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Molecular Ecology 10:249–256. 10.1046/j.1365-294X.2001.01185.x [DOI] [PubMed] [Google Scholar]
- Walker JB, Sytsma KJ, Treutlein J, Wink M. 2004. Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. American Journal of Botany 91:1115–1125. 10.3732/ajb.91.7.1115 [DOI] [PubMed] [Google Scholar]
- Welch DM, Meselson M. 2000. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 288:1211–1215. 10.1126/science.288.5469.1211 [DOI] [PubMed] [Google Scholar]
- Zeder MA. 2008. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proceedings of the National Academy of Sciences of the USA 105:11597–11604. 10.1073/pnas.0801317105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder MA, Hesse B. 2000. The initial domestication of goats (Capra hircus) in the Zagros Mountains 10,000 years ago. Science 287:2254–2257. 10.1126/science.287.5461.2254 [DOI] [PubMed] [Google Scholar]