Table 2.
Proposed neutral and acidic oligosaccharide structures or sequences identified in human mucins from tumors and resection margins of patients with colorectal cancers and from colonic mucosa of healthy individuals.
| Proposed structures or sequences of oligosaccharides | [M + Na]+ or [M + 2Na-H]+ for sulfated species | Tumors | Resection margins | Healthy controls |
|---|---|---|---|---|
 |
534 | 5.6 ± 5.7 | 1.8 ± 2.1 | 1.0 ± 1.4 |
 |
575 | 1.7 ± 1.5 | 5.8 ± 6.7 | 3.8 ± 2.5 |
 |
663 | 0.2 ± 0.4 | 0.3 ± 0.6 | 1.0 ± 1.4 |
 |
691 | 7.1 ± 7.1 | 5.2 ± 5.3 | 5.8 ± 4.9 |
 |
708 | 0.5 ± 1 | 0.3 ± 0.6 | 0.4 ± 0.5 |
 |
749 | 0.4 ± 0.8 | 1.0 ± 2.2 | 0 |
|
779 | 4.2 ± 1.5 | 5.0 ± 3.8 | 5.6 ± 0.4 |
 |
867 | 0.8 ± 1.1 | 0.3 ± 0.6 | 0 |
|
895 | 10.9 ± 3.6 | 7.7 ± 4.2 | 5.7 ± 0.6 |
 |
908 | 0.6 ± 1.0 | 1.3 ± 1.8 | 0.8 ± 1.1 |
 |
936 | 12.1 ± 11.2 | 24.2 ± 10.2 | 30.8 ± 11.6 |
 |
953 | 1.4 ± 0.6 | 3.0 ± 1.8 | 1.9 ± 0.7 |
 |
983 | 7.5 ± 8.3 | 2.0 ± 0.7 | 1.6 ± 0.3 |
 |
1,024 | 0.2 ± 0.4 | 0.4 ± 0.5 | 0.3 ± 0.5 |
 |
1,041 | 0.0 ± 0.1 | 0.3 ± 0.6 | 0.2 ± 0.2 |
 |
1,069 | 0.2 ± 0.4 | 0.2 ± 0.4 | 0.8 ± 0.1 |
 |
1,071 | 0.6 ± 1.5 | 0 | 0 |
 |
1,110 | 0.3 ± 0.5 | 0.7 ± 1.1 | 0.7 |
 |
1,112 | 0.1 ± 0.2 | 0.2 ± 0.3 | 0.3 ± 0.4 |
 |
1,127 | 0 | 0.3 ± 0.8 | 0 |
 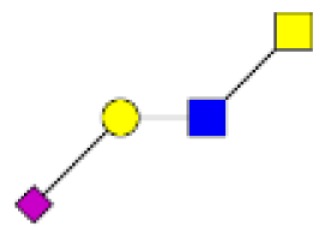
|
1,140 | 2.9 ± 2.8 | 4.4 ± 1.9 | 7.0 |
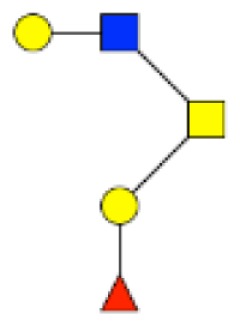 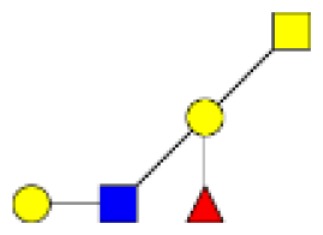
|
1,157 | 1.9 ± 1.2 | 0.4 ± 0.7 | 0.3 ± 0.4 |
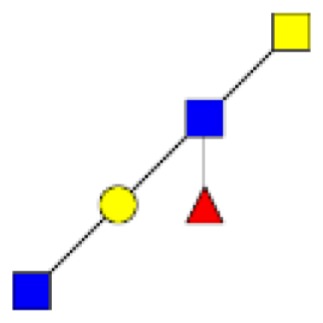 |
1,198 | 0.0 ± 0.1 | 0.4 ± 0.6 | 0 |
 |
1,228 | 0.8 ± 0.5 | 0.5 ± 0.5 | 0.6 ± 0.2 |
 |
1,256 | 3.5 ± 1.8 | 2.7 ± 4.0 | 1.5 ± 0.4 |
|
1,314 | 3.3 ± 2.4 | 7.0 ± 4.3 | 8.0 ± 1.4 |
 |
1,331 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.1 ± 0.2 |
 |
1,344 | 7.5 ± 5.5 | 3.3 ± 6.0 | 1.1 ± 0.4 |
 |
1,385 | 0.7 ± 1.4 | 1.9 ± 2.3 | 0.7 ± 1.0 |
 |
1,402 | 0.8 ± 0.6 | 3.2 ± 1.4 | 7.1 ± 0.3 |
| 3 Hex, 2 HexNAc, GalNAcol | 1,432 | 0.9 ± 0.8 | 0.2 ± 0.2 | 0.2 ± 0.3 |
| 2 Hex, 3 HexNAc, GalNAcol | 1,473 | 0.1 ± 0.2 | 0.1 ± 0.2 | 0.2 ± 0.3 |
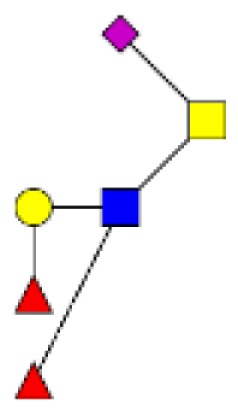 |
1,488 | 0.3 ± 0.6 | 0.8 ± 1.7 | 0.7 ± 1.0 |
| 2 Hex, 2 HexNAc, 1 Fuc, 1 SO3, GalNAcol | 1,490 | 0.2 ± 0.4 | 0 | 0 |
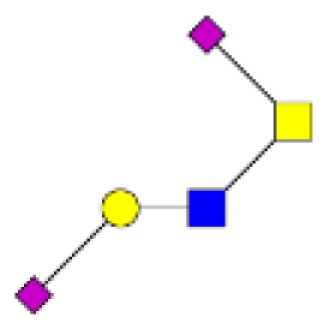 |
1,501 | 0.5 ± 0.6 | 0.6 ± 0.9 | 1.6 ± 0.7 |
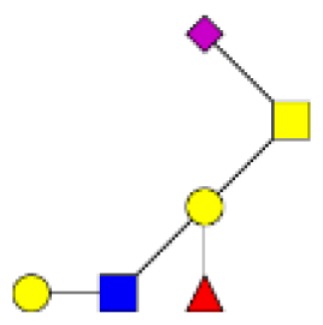 |
1,518 | 1.7 ± 1.8 | 0.4 ± 0.6 | 0.2 ± 0.4 |
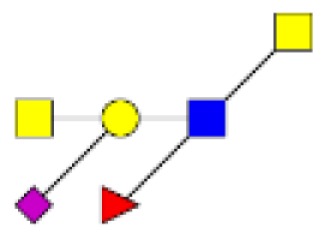 |
1,559 | 0.1 ± 0.2 | 0.1 ± 0.2 | 0.1 ± 0.2 |
| 2 Hex, 2 HexNAc, 2 Fuc, GalNAcol | 1,576 | 0 ± 0.1 | 0 | 0.1 ± 0.2 |
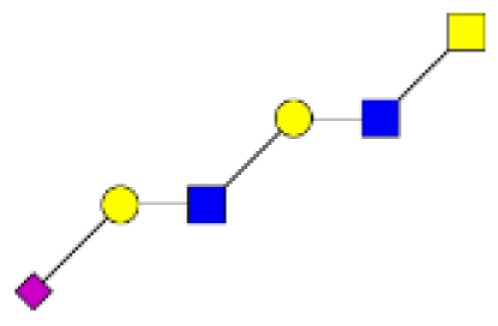 |
1,589 | 0.3 ± 0.3 | 0.3 ± 0.5 | 0.4 ± 0.1 |
| 3 Hex, 2 HexNAc, 1 Fuc, GalNAcol | 1,606 | 0.7 ± 0.8 | 0 | 0.1 ± 0.1 |
| 2 Hex, 3 HexNAc, 1 Fuc, GalNAcol | 1,647 | 0.6 ± 0.8 | 0.2 ± 0.4 | 0.3 |
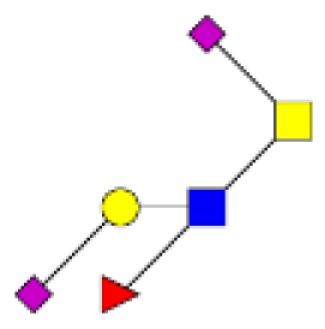 |
1,675 | 9.4 ± 1.2 | 2.5 ± 2.4 | 3.3 ± 0.7 |
| 3 Hex, 3 HexNAc, GalNAcol | 1,677 | 1.0 ± 1.3 | 0.4 ± 1 | 0 |
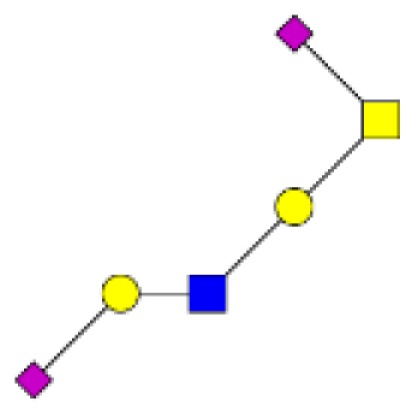 |
1,705 | 3.2 ± 3.4 | 2.5 ± 6.0 | 0.6 ± 0.1 |
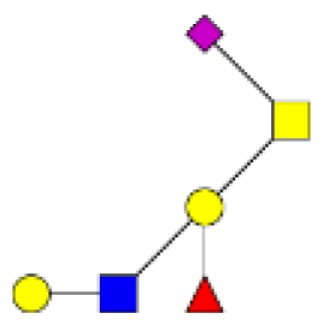 |
1,718 | 0.2 ± 0.3 | 0.1 ± 0.3 | 0.1 ± 0.1 |
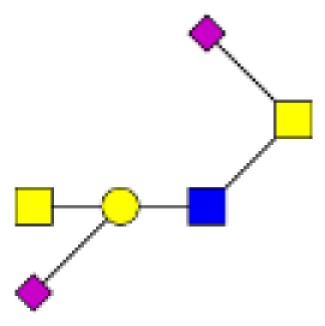 |
1,746 | 0.9 ± 2 | 1.9 ± 2.3 | 1.1 ± 1.5 |
| 2 Hex, 2 HexNAc, 1 Fuc, 1 NeuAc, GalNAcol | 1,763 | 0.2 ± 0.2 | 0.2 ± 0.3 | 0.3 ± 0.1 |
| 3 Hex, 2 HexNAc, 2 Fuc, GalNAcol | 1,780 | 0.5 ± 0.8 | 0 | 0 |
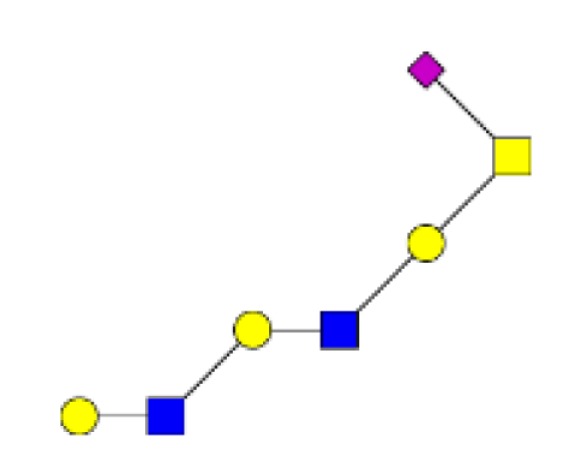 |
1,793 | 0.3 ± 0.4 | 0.2 ± 0.4 | 0 |
| 2 Hex, 3 HexNAc, 1 NeuAc, GalNAcol | 1,834 | 0.1 ± 0.1 | 0.2 ± 0.3 | 0.1 ± 0.1 |
| 3 Hex, 3 HexNAc, 1 Fuc, GalNAcol | 1,851 | 0.4 ± 0.8 | 0 | 0.1 ± 0.1 |
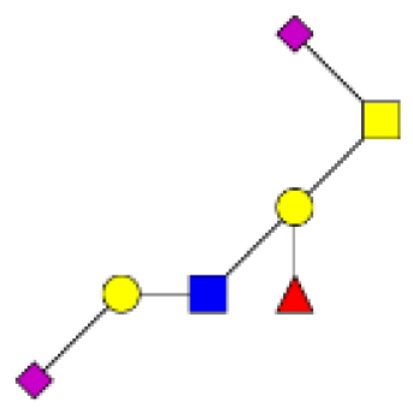 |
1,879 | 0.2 ± 0.5 | 0.3 ± 0.8 | 0 |
| 4 Hex, 3 HexNAc, GalNAcol | 1,881 | 0.4 ± 0.7 | 0 | 0 |
| 4 Hex, 2 HexNAc, 1 Fuc, 1 SO3, GalNAcol | 1,898 | 0 ± 0.1 | 0.1 ± 0.2 | 0 |
| 3 Hex, 4 HexNAc, GalNAcol | 1,922 | 0.2 ± 0.5 | 0.1 ± 0.3 | 0.1 ± 0.1 |
| 2 Hex, 2 HexNAc, 2 Fuc, 1 NeuAc, GalNAcol | 1,937 | 0 ± 0.1 | 0 | 0.1 ± 0.1 |
| 3 Hex, 3 HexNAc, 1 Fuc, 1 SO3, GalNAcol | 1,939 | 0.1 ± 0.2 | 0 | 0 |
| 3 Hex, 2 HexNAc, 3 Fuc, GalNAcol | 1,954 | 0.1 ± 0.2 | 0 | 0 |
| 4 Hex, 2 HexNAc, 1 NeuAc, GalNAcol | 1,997 | 0.1 ± 0.2 | 0 | 0 |
| 3 Hex, 3 HexNAc, 2 Fuc, GalNAcol | 2,025 | 0.4 ± 0.5 | 0.1 ± 0.2 | 0.1 ± 0.1 |
| 5 Hex, 3 HexNAc, GalNAcol | 2,055 | 0.6 ± 0.9 | 0 | 0 |
The relative percentage of each oligosaccharide was calculated based on the integration of peaks on MS spectra. Results are presented as the mean ± SD of percentage of each oligosaccharide for a same condition.
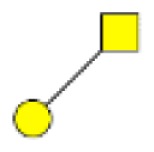
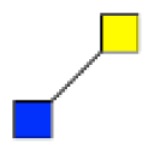
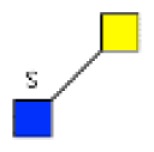
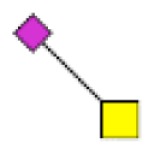
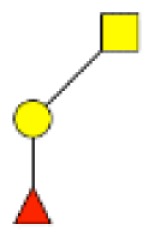
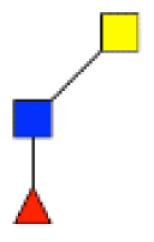
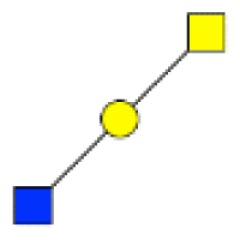
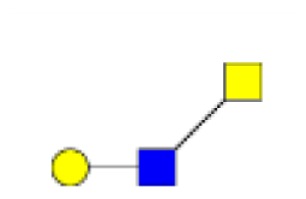
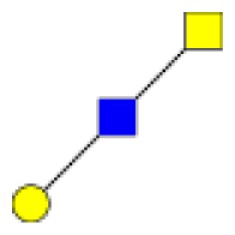
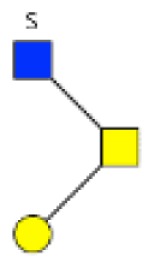
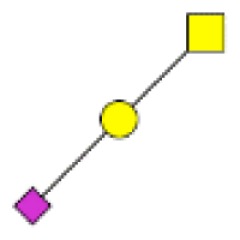
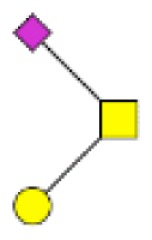
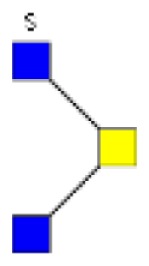
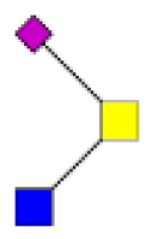
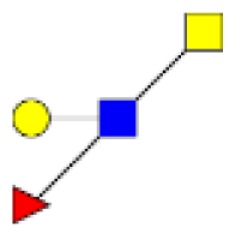
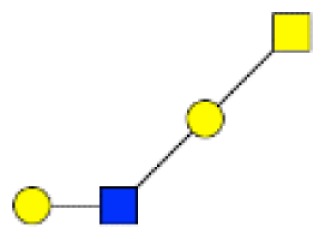
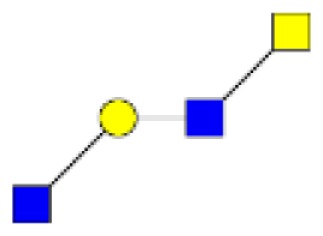
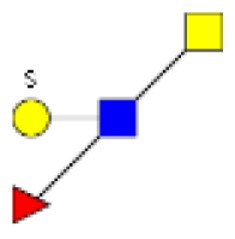
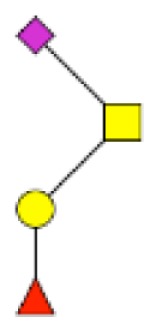
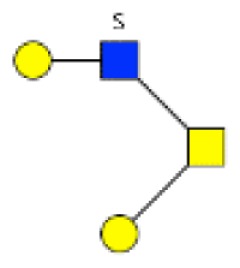
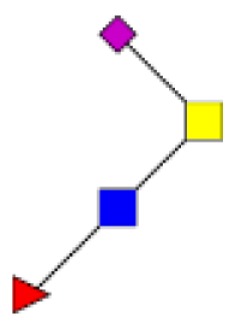
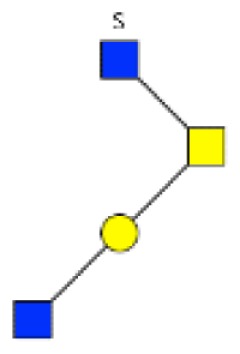
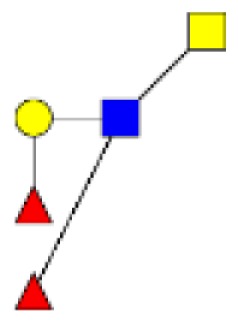
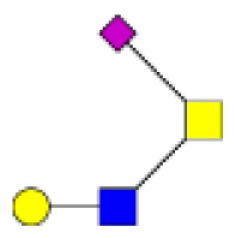
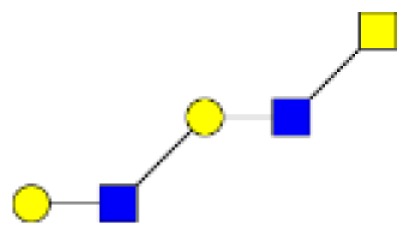
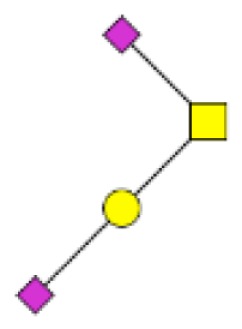
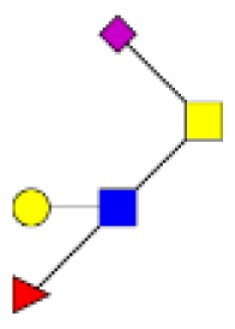
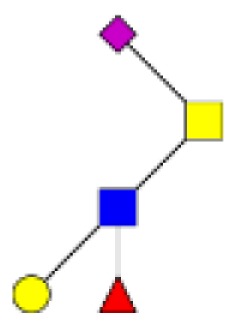
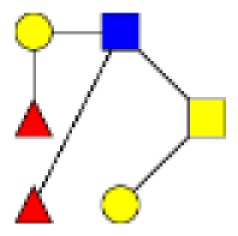
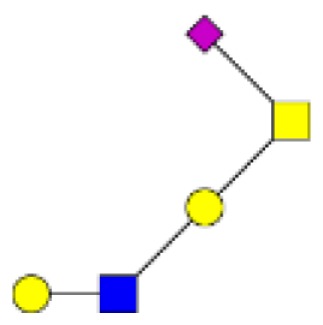
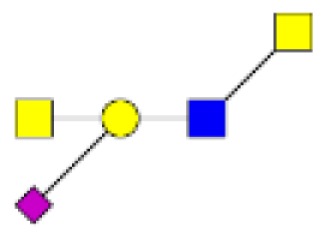
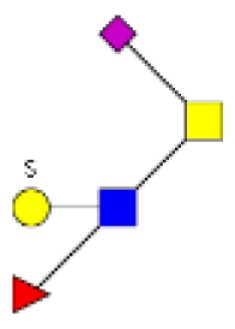
Monosaccharide symbols according to the Consortium for Functional Glycomics (CFG) nomenclature. Key: fucose (red triangle), GlcNAc (blue square), sialic acid (purple diamond), galactose (yellow circle), GalNAcol (yellow square), and sulfate residue (S).
