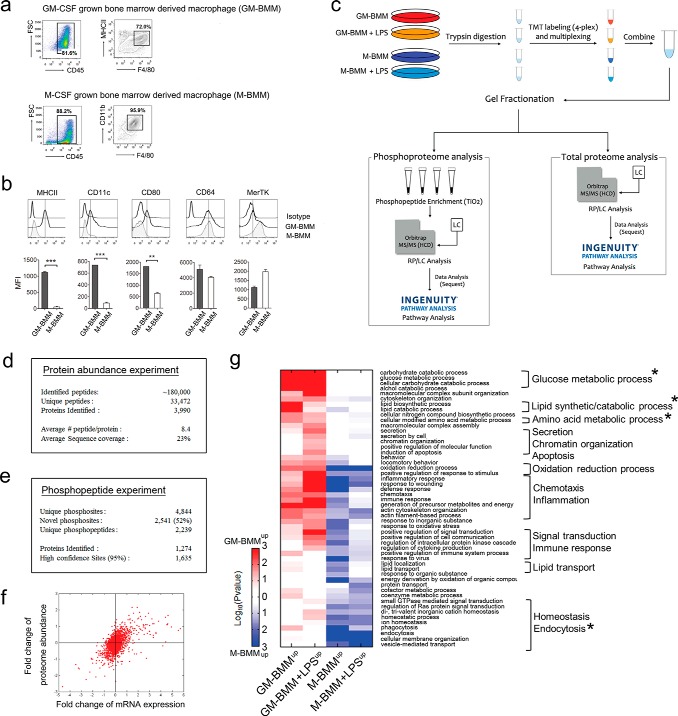
Fig. 1.
Quantitative proteomic/phosphoproteomic analysis of GM-BMMs/M-BMMs. A, GM-BMMs and M-BMMs were differentiated over 7 days from C57BL/6J male mice bone marrow cells with GM-CSF or M-CSF supplementation, respectively. Cells were sorted by CD45+F4/80+MHCII+ population for GM-BMMs, or CD45+F4/80+CD11b+ population for M-BMMs. B, Surface marker examination of GM-BMMs and M-BMMs. Attached cells were stained with the appropriate antibodies and analyzed for surface MHCII, CD11c, CD80, CD64, and MerTK expressions per gated populations in A. Three independent experiments were performed. **, p < 0.01, ***, p < 0.001 by Student t test. C, Experimental strategy. Macrophage proteins were pooled to generate three biological replicates for each of the GM-BMM/M-BMM/GM-BMM+LPS/M-BMM+LPS experimental groups. D, Results of protein expression experiments. E, Results of phosphoproteome experiments. F, Scatter plot of gene symbols detected both in proteome and microarray analyses. Data represents the fold change of log2(GM-BMM/M-BMM) of 2491 genes. Regression p value = 2.1987e-242, R-squre = 0.31352. G, GO analysis of differentially expressed proteins and phosphopeptides. Heat-map shows significant GO biological process terms (p < 0.05) for differentially expressed proteins/phosphoproteins between GM-BMM/M-BMM and GM-BMM+LPS/M-BMM+LPS. Red color indicates increased protein/phosphoprotein abundances in GM-BMM or GM-BMM/LPS, and blue color indicates increased abundances in M-BMM or M-BMM/LPS. Star markings show the most enriched GO category in a heatmap.