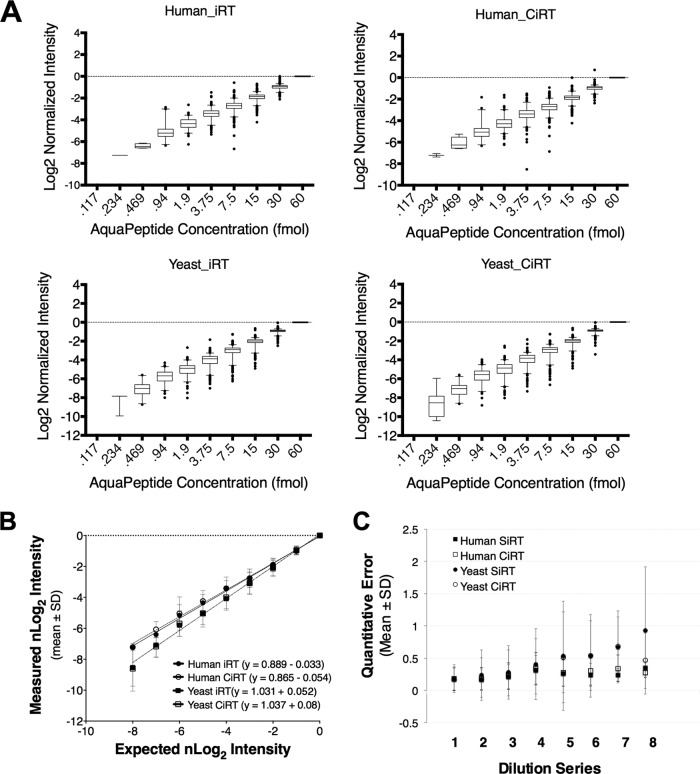
Fig. 5.
Use of internal retention time prediction peptides does not alter the accuracy of peptide quantitation by SWATH-MS. A, Synthetic, heavy peptides were spiked into cell lysate from either human-derived (upper panel) or yeast (lower panel) cells at progressively twofold decreasing concentrations from 30 femtomoles to .058 femtomoles on column. Assay libraries, normalized to iRT, for the 340 synthetic peptides were used to extract peak groups from SWATH files, and the intensity of a given peak group was normalized to its observed intensity at 30 fmol and set to log scale with a base of 2. Perfectly accurate quantification would therefore represent a single unit increase between dilution steps. Quantification subsequent to synthetic iRT prediction is shown on the left and the CiRT shown on the right. B, Comparison of the linear estimate comparing the mean observed normalized Log2 Intensities plotted against that which would be expected based on the actual concentration of heavy peptide spiked into a given sample. Each data point represents the mean ± standard deviation (S.D.) of nLog2 intensity across all peptides observed at a given concentration, C, A plot of the mean absolute error calculated for each dilution step of the 10 × 2-fold dilution series (described in methods). No peptides were detected in the lowest concentration, and as such there are 8 dilution steps used to calculate quantification error see methods for the equation used for error estimation). Values are presented as mean ± S.D. at each dilution step, for each RT normalization method (CiRT versus SiRT) in Yeast and Human samples.