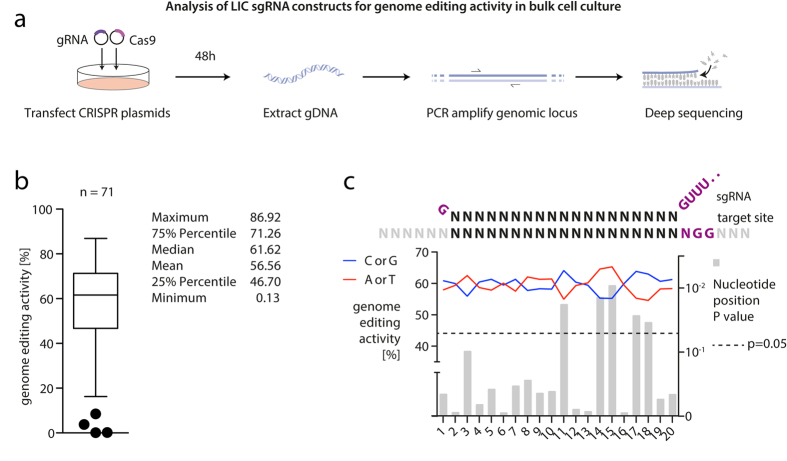
Figure 2. Analysis of LIC sgRNA constructs for genome editing activity.
(a) sgRNA plasmids were generated by LIC and co-transfected with a Cas9 expression plasmid into HEK 293T cells. After 48 h cells were lysed, the targeted loci amplified by PCR, and the products analyzed using deep sequencing. (b) Genome editing activity of 71 sgRNAs is summarized in a box plot, whereas Tukey outliers are depicted as individual dots. (c) Shown is the mean genome editing activity of 67 CRISPR constructs (all CRIPSR, except the 4 outliers) calculated from the activity of all sgRNAs that have either a G/C or A/T at the indicated nucleotide position. The most 5′ base of the target site was determined as position 1, whereas the position 21 is the base N of the PAM (NGG). PAM is depicted in purple. Statistical analysis was carried out as an unpaired t-test, whereas the calculated p-values are depicted as bar charts. The dashed black line highlights the p-value threshold of 0.05.