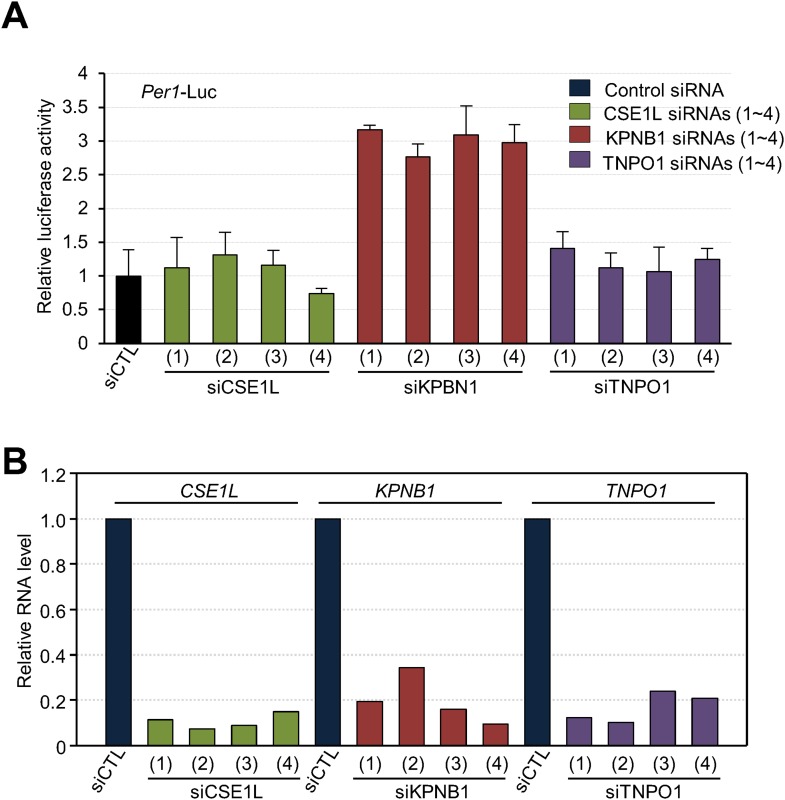
Figure 3. KPNB1 mediates PERs/CRYs-regulated transcription of E-box dependent clock genes.
(A) HEK293T cells were transiently transfected with Per1-Luc reporter construct alone or cotransfected with plasmids expressing CLOCK, BMAL1, PER1, PER2, CRY1, CRY2, in the presence of control (siCTL: black), CSE1L (siCSE1L: light green), KPNB1 (siKPNB1; gray red), and TNPO1 siRNA (siTNPO1: violet) as indicated. After 24 hr, the cells were lysed and Per1 promoter-driven luciferase activities were measured and normalized with pRL-TK activity. Results of one representative experiment of three independent experiments are shown. (B) Schematic diagram of the human PER1 promoter and primers used for ChIP assay. (C, D) ChIP assay of PER1 or PER2 binding to the E box in hPER1. Control (siCTL: black) and KPNB1 siRNA (siKPNB1; gray red)-treated cells (U2 OS) were subjected to ChIP assays using anti-PER1 (αPER1) or anti-PER2 (αPER2) antibody. ChIP DNA samples were quantified by quantitative real-time RT-PCR. The data presented are the means ± S.E. of triplicate samples (**p < 0.005, *p < 0.05, by Student's t-test). (E) Quantitative real-time RT-PCR analysis of expression of endogenous PER1, CRY1, DBP, REVERBβ, and BMAL1 mRNAs in control (siCTL: black), KPNB1-depleted (siKPNB1; gray red) or overexpressed (KPNB1OV; blue green) cells (U2 OS). The data presented are the means ± S.E. of triplicate samples (**p < 0.005, *p < 0.05, by Student's t-test).