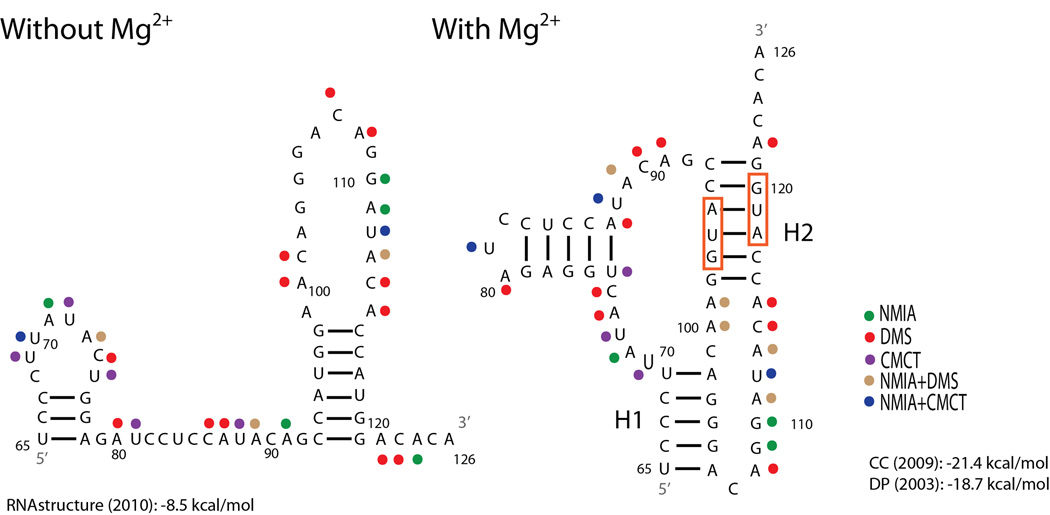
Figure 1.
Structural models of the influenza A segment 2 (PB1) 65–126 nucleotide region. To optimize SP6 polymerase efficiency the identities of nucleotides 63 and 64 (not shown) are both A’s, however in the wild-type sequence the nucleotides are A and U, respectively. The structure without Mg2+ is predicted with RNA structure15 where strongly reactive nucleotides were forced to be single-stranded. The structure shown in the presence of Mg2+ is the one most consistent with the model based on sequence comparison.16 This structure is also predicted by ShapeKnots.17 Predicted free energy at 37 °C for the structure without Mg2+ is reported at the bottom left.15 Predicted free energies at 37 °C from two pseudoknot prediction models are reported at the bottom right.18,19 Red boxes indicate the start codons for PB1-F2 and N40 ORFs, respectively. Colored dots represent strong chemical mapping hits for the reagent identified in the key. H1: Helix 1, H2: Helix 2.