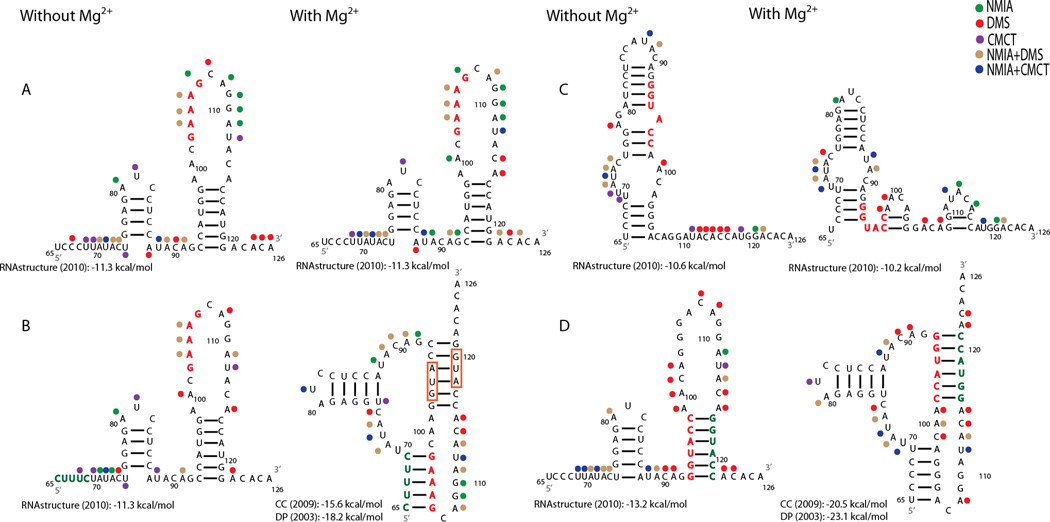
Figure 2.
Chemical mapping results for four mutant sequences with and without Mg2+. Red nucleotides indicate predicted destabilizing nucleotide changes and green represents compensatory changes. Predicted non-pseudoknotted structures were modeled with RNA structure as in Figure 1. Predicted pseudoknotted structures (B and D with Mg2+) were previously modeled with ShapeKnots and overlaid with chemical mapping results. (A) Destabilizing mutant sequence for Helix 1. (B) Compensatory mutant sequence for Helix 1. (C) Destabilizing mutant sequence for Helix 2. (D) Compensatory mutant sequence for Helix 2. Other figure annotations are as in Figure 1.