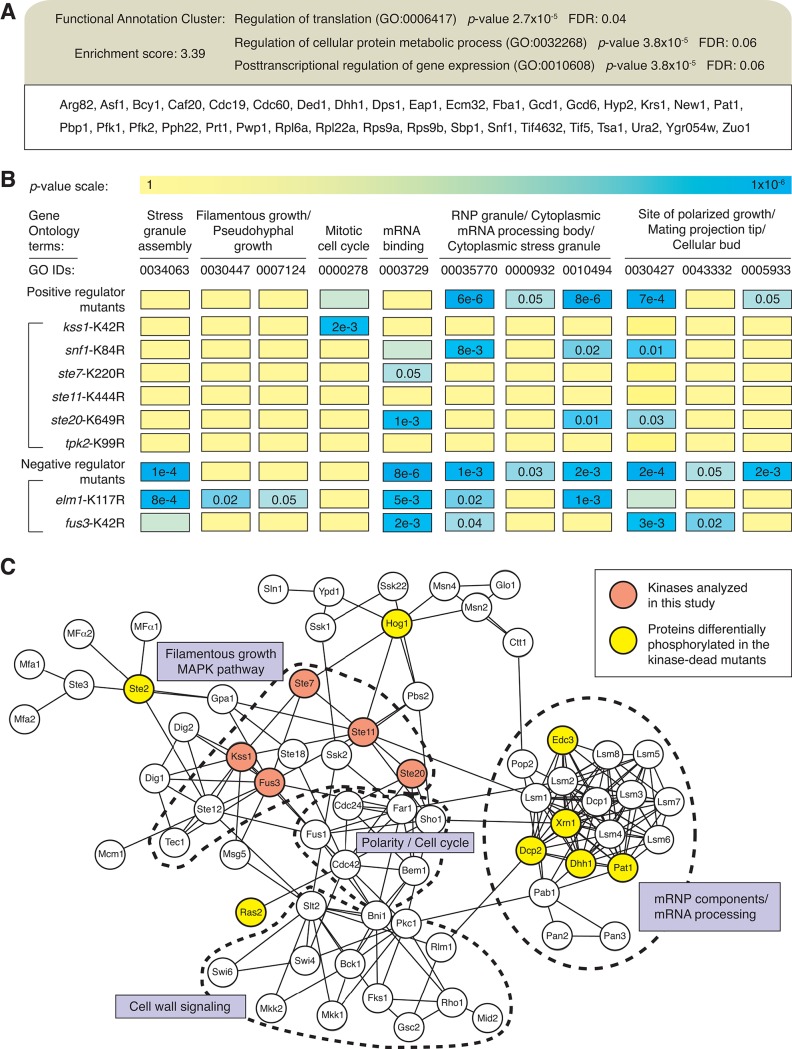
Fig 3. The yeast pseudohyphal growth kinase signaling network is enriched in proteins contributing to mRNP granule function.
A) Analysis of phosphoproteins identified as being dependent upon yeast pseudohyphal growth kinases indicates a cluster of proteins involved in translational regulation, encompassing protein components of RNPs. B) The diagram presents a heat map for the enrichment of indicated GO terms (columns) in the respective mass spectrometry data sets generated from the kinase mutants (rows). For convenience, GO IDs are listed below each GO term. Kinases are grouped as positive and negative regulators and presented as individual data sets. C) Connectivity map of proteins in KEGG pathways associated with pseudohyphal growth (MAPK signaling pathway sce04011, cell cycle sce04111, meiosis sce04113) and proteins identified as being components of either P-bodies or stress granules. Lines indicate interactions annotated in KEGG and iRefIndex. Dashed lines identify clusters of proteins involved in the indicated processes.