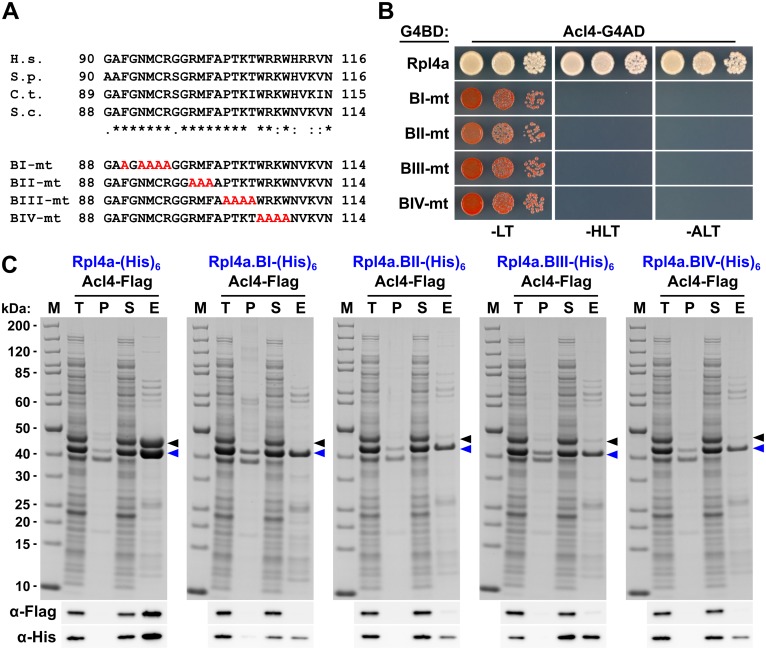
Fig 6. Multiple Rpl4 residues are required for the interaction with Acl4.
A, Amino acid sequences of the minimal Acl4 binding site within the C-terminal part of the long internal loop of Rpl4 from different eukaryotic species (H.s., Homo sapiens; S.p., Schizosaccharomyces pombe; C.t., C. thermophilum; S.c., S. cerevisiae). Conserved (*), strongly similar (:), and weakly similar (.) amino acids are indicated below the alignment. The non-overlapping, consecutive alanine substitutions within this Rpl4a segment are depicted in the lower part: block-I mutant (BI-mt): F90A/N92A/M93A/C94A/R95A, block-II mutant (BII-mt): R98A/M99A/F100A, block-III mutant (BIII-mt: P102A/T103A/K104A/T105A, and block-IV mutant (BIV-mt): W106A/R107A/K108A/W109A). B, Y2H interaction between the Rpl4 alanine block mutants and Acl4. Plasmids expressing full-length Rpl4a or the indicated Rpl4a mutant variants, fused to the C-terminal Gal4 DNA-binding domain (G4BD), and full-length Acl4, fused to the C-terminal Gal4 activation domain (G4AD), were co-transformed into the Y2H reporter strain PJ69-4A. Cells were spotted in 10-fold serial dilution steps onto SC-Leu-Trp (-LT), SC-His-Leu-Trp (-HLT), and SC-Ade-Leu-Trp (-ALT) plates, which were incubated for 3 d at 30°C. C, In vitro interaction between the Rpl4 alanine block mutants and Acl4. Full-length Rpl4a or the indicated Rpl4a variants, containing a C-terminal (His)6 tag, were co-expressed with full-length Acl4-Flag in E. coli and purified via Ni-affinity purification. Proteins were revealed by SDS-PAGE and Coomassie staining (top) or by Western blot analysis using anti-Flag (Acl4-Flag) and anti-His (Rpl4a-(His)6 variants) antibodies (bottom). T, total extract; P, pellet fraction (insoluble proteins); S, soluble extract; E, imidazole eluate; M, molecular weight standard. Blue arrowheads highlight the bands corresponding to the different Rpl4a-(His)6 variants used as baits for the purifications. Black arrowheads indicate the position of Acl4-Flag.