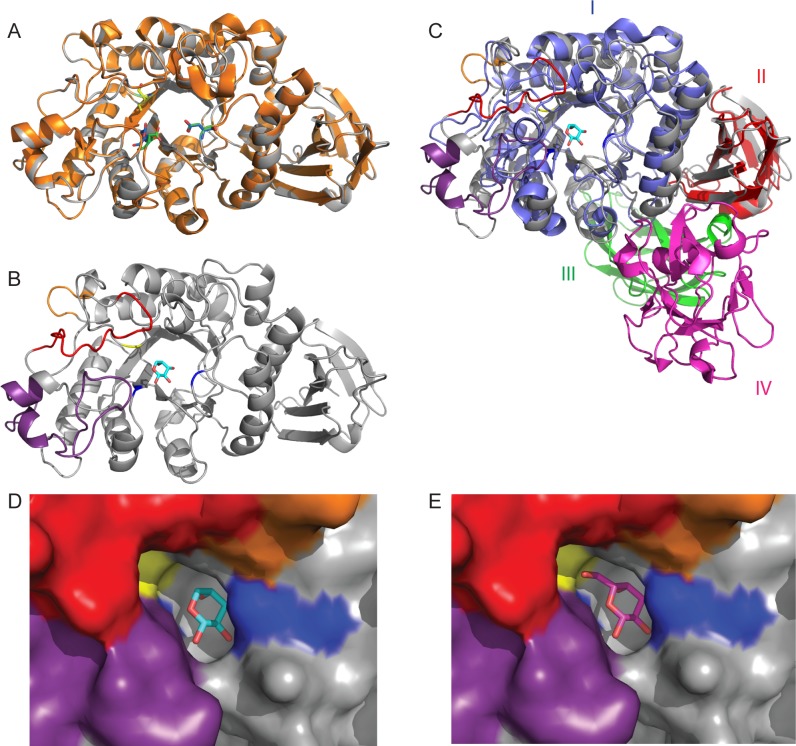
Fig 6. Structural comparison of CpArap27 with GsAbp and SaArap27A.
(A) Cartoon representation of the CpArap27 homology model (grey) and the crystal structure of GsAbp (orange). Catalytic amino acids for the enzymes are picked out respectively in blue/green, while the key Isoleucine specificity determinant is yellow/cyan. (B) Secondary structural representation of CpArap27, with loop insertions highlighted as follows: L6 is red, L7 is orange, and L8 is purple. Ile56 is shown in stick form and highlighted in yellow. Catalytic amino acids are highlighted in blue. (C) Comparison of the modular structures of SaArap27A (Domains I-IV) and the homology model of CpArap27 (Domains I and II). SaArap27A is shown in 4 colours to highlight the 4 domains of this protein. CpArap27 is coloured as in panel B. A molecule of l-Ara is shown in the active site, taken from the structure of SaArap27A. (D) and (E) Surface representations of the active site of the CpArap27 homology model overlaid with, respectively, l-Ara and d-Gal from the SaArap27A structures. Catalytic amino acids are shown in blue, Ile56 is shown in yellow, and the loop insertions are again shown in purple, red and orange. The Isoleucine shown here is predicted to hydrophobically clash with a hypothetical d-Gal substrate, as shown for GsAbp [51].