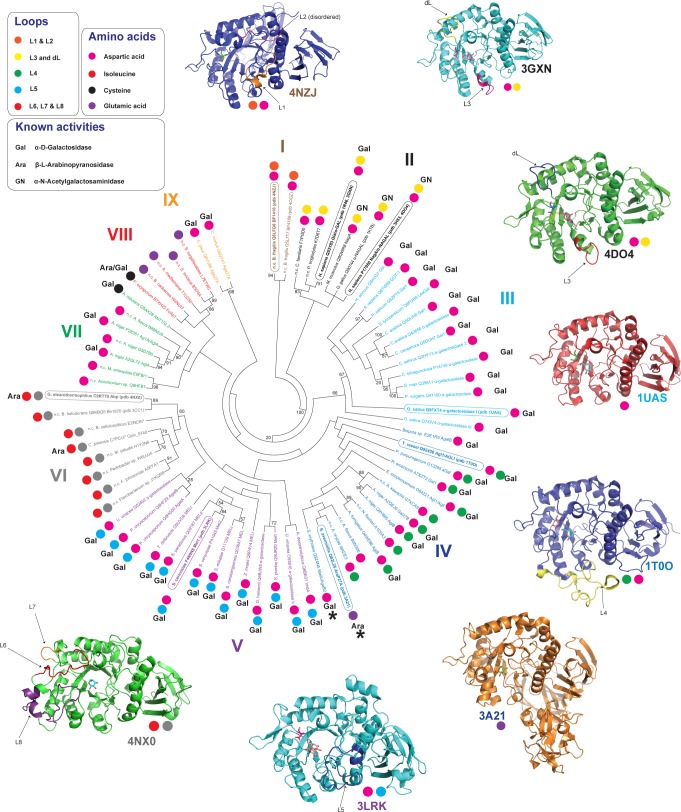
Fig 7. Phylogenetic analysis of family GH27.
Proteins are labelled with organism name and UNIPROT code. The phylogenetic tree includes all characterised GH27 enzymes identified as such on the CAZy database at the time of writing, plus several as yet uncharacterised gene products, identified by sequence homology to characterised enzymes using pBLAST analysis. Proteins for which a 3D structure is available are shown in bold and pdb codes are provided. Examples are shown in cartoon form around the tree to highlight conserved structural elements within a clade, and the name of an illustrated protein is circled on the tree. Each clade is labelled in a specific colour. Characterised enzymes are labelled with Ara or Gal to indicate their preferred substrate. Currently uncharacterised proteins are indicated with n.c.. Finally, proteins not conforming to the pattern of loop insertions or active site amino acids otherwise well conserved for their clade are highlighted with an asterisk. The phylogenetic tree was produced using a Clustal Omega alignment [31] and the PhyML software [32], and the tree was visualised using MEGA5 [67].