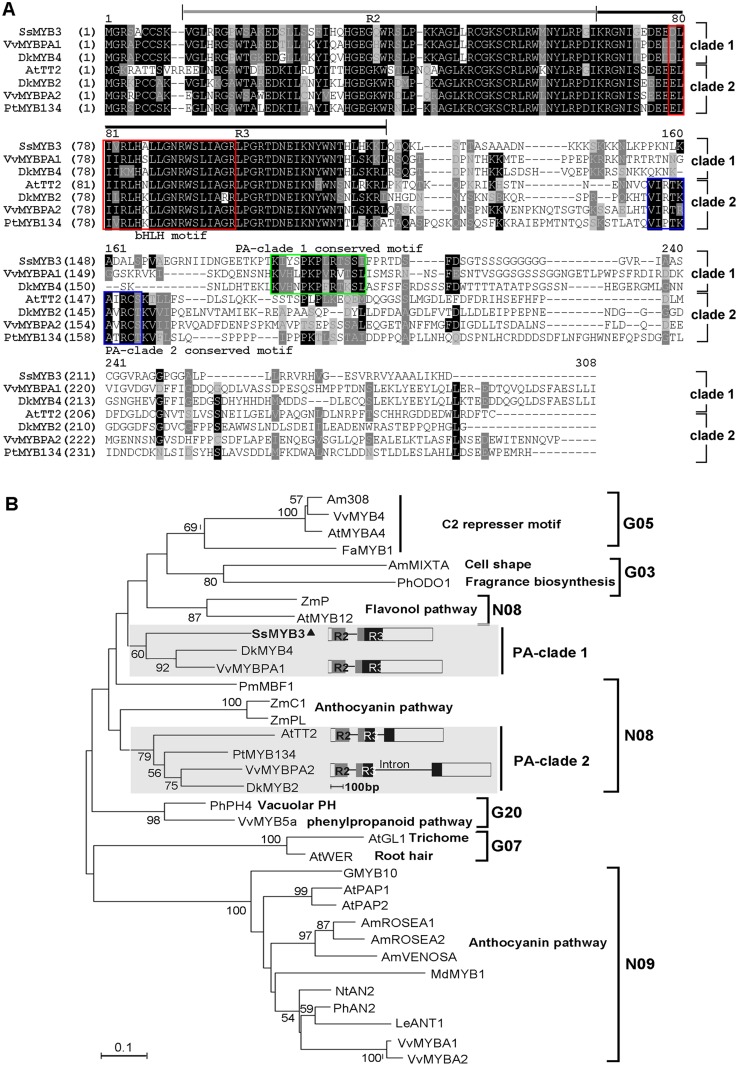
Fig 2. Comparison of SsMYB3 with other functional R2R3-MYB TFs.
(A) Full-length protein sequence alignments of the MYBs by Vector NTI 10.0 software. The R2 and R3 domains are shown; the bHLH motif [D/E]Lx2[R/K]x3Lx6Lx3R and two possible PA specifically conserved motifs, K[I/V]x2PKPx1Rx2S[I/L] and VI[R/P]TKAx1RC[S/T], are indicated in the green and blue box, respectively. (B) Phylogenetic tree analysis depicts SsMYB3 belonging to PA-clade 1. The tree was constructed from the ClustalX 1.8 alignment using the neighbor-joining method in the MEGA 5.0 program. Scale bar represents 0.1 substitutions per site, and numbers next to nodes are bootstrap values from 1,000 replicates (only values >50% are shown). SsMYB3 and other known functional PA-biosynthesis regulators are divided into two subclades (PA-clades 1 and 2) with different genomic structures indicated in gray background. MYB proteins with known functions and MYB subgroups (G03, G05, G07, G20, N08, and N09) are indicated. GenBank accession numbers of MYBs in the phylogenetic tree are as follows: AmMIXTA (CAA55725), AmROSEA1 (ABB83826), AmROSEA2 (ABB83827), AmVENOSA (ABB83828), Am308 (JQ0960), AtTT2 (Q9FJA2), AtPAP1 (ABB03879), AtPAP2 (NP_176813), AtMYB4 (BAA21619), AtMYB12 (ABB03913), AtWER (AAF18939), AtGL1 (AAC97387), DkMYB2 (AB503699), DkMYB4 (AB503671), FaMYB1 (AAK84064), GMYB10 (CAD87010), LeANT1 (AAQ55181), MdMYB1 (DQ886414), PmMBF1 (AAA82943), PhAN2 (AAF66727), PhPH4 (AAY51377.1), PhODO1 (AAV98200), PtMYB134 (ACR83705), ZmC1 (AAA33482), ZmPl (AAA19821), ZmP (AAC49394), VvMybPA1 (CAJ90831), VvMybPA2 (EU919682), VvMYBPA1 (BAD18977), VvMYBPA2 (BAD18978), VvMYB4 (ABL61515), and VvMYB5a (AAS68190).