Abstract
Advanced techniques including the chromosome conformation capture (3C) methodology and its derivatives are complementing microscopy approaches to study genome organization, and are revealing new details of three-dimensional (3D) genome architecture at increasing resolution. The fission yeast Schizosaccharomyces pombe (S. pombe) comprises a small genome featuring organizational elements of more complex eukaryotic systems, including conserved heterochromatin assembly machinery. Here we review key insights into genome organization revealed in this model system through a variety of techniques. We discuss the predominant role of Rabl-like configuration for interphase chromosome organization and the dynamic changes that occur during mitosis and meiosis. High resolution Hi-C studies have also revealed the presence of locally crumpled chromatin regions called “globules” along chromosome arms, and implicated a critical role for pericentromeric heterochromatin in imposing fundamental constraints on the genome to maintain chromosome territoriality and stability. These findings have shed new light on the connections between genome organization and function. It is likely that insights gained from the S. pombe system will also broadly apply to higher eukaryotes.
Keywords: heterochromatin, cohesin, Rabl, genome organization, Hi-C, fission yeast
1. Introduction
Determining how chromosomes, which contain the genetic information specifying proper developmental and gene expression programs, are organized within the nuclear space has remained a major driving force in the nuclear architecture field [1–3]. The physical compaction of chromosomes and the spatial organization of the genome are critical for maintaining genome stability and for the proper regulation of many nuclear functions, including transcription, replication, recombination, and repair [4–7]. Several layers of organization are imposed on the chromatin fiber, ranging from nucleosomal packaging to intricate levels of higher-order folding. Factors involved in chromatin assembly, including heterochromatin machinery that targets specific genomic sites and architectural proteins such as condensin and cohesin, play important roles in genome organization. Exactly how these factors contribute to the organization of genome function and how they facilitate dynamic changes in chromosome architecture during the cell cycle are just beginning to be revealed.
The advent of several advanced techniques has allowed packaging, folding, and genomic interactions to be studied in increasingly fine detail. Molecular based approaches, including 3C and its derivatives, have been used to study specific loci of interest as well as genome-wide interactions to gain insight into the physical organization and spatial configuration of chromosomes [8–13]. Short and long-range interactions both within and between chromosomes have been determined from global interaction contact maps obtained by these methods. These studies have yielded important conceptual advances, including the compartmentalization of the human genome into open and closed states, chromosome territoriality, and the fractal globule nature of the chromatin fiber [13]. Evidence of megabase-sized topologically associating domains (TADs) has been discovered in various systems [14–16]. In parallel, advanced microscopy studies at increasing resolution are complementing molecular studies, providing visual clues to genome organization at single cell resolution [17,18].
Among the various model organisms, the fission yeast S. pombe has emerged as a useful system to study 3D genome organization. S. pombe comprises a small genome with hallmarks of more complex eukaryotes. The 13.8 Mb S. pombe genome is comprised of three relatively large chromosomes. Centromeres ranging in size from ~35–110 kb are organized into two distinct domains: the unique central core bound by CENP-A and kinetochore proteins, and the surrounding pericentromeric repeats [19,20] (Figure 1A). Extended heterochromatin domains coat pericentromeric repeats and subtelomeric regions as well as the silent mating-type (mat) interval (Figure 1A) [21]. Studies of heterochromatin assembly pathways involving conserved proteins, such as Clr4/Suv39h and Swi6/HP1 that are present in S. pombe, have provided insights into the critical functions of this specialized chromatin [20,22,23]. In particular, work that focused on the mat locus has yielded many groundbreaking discoveries over the years, including the epigenetic inheritance of differential chromatin states and the mechanisms by which boundary DNA elements prevent spreading of heterochromatin into neighboring gene-rich euchromatin regions [24–27].
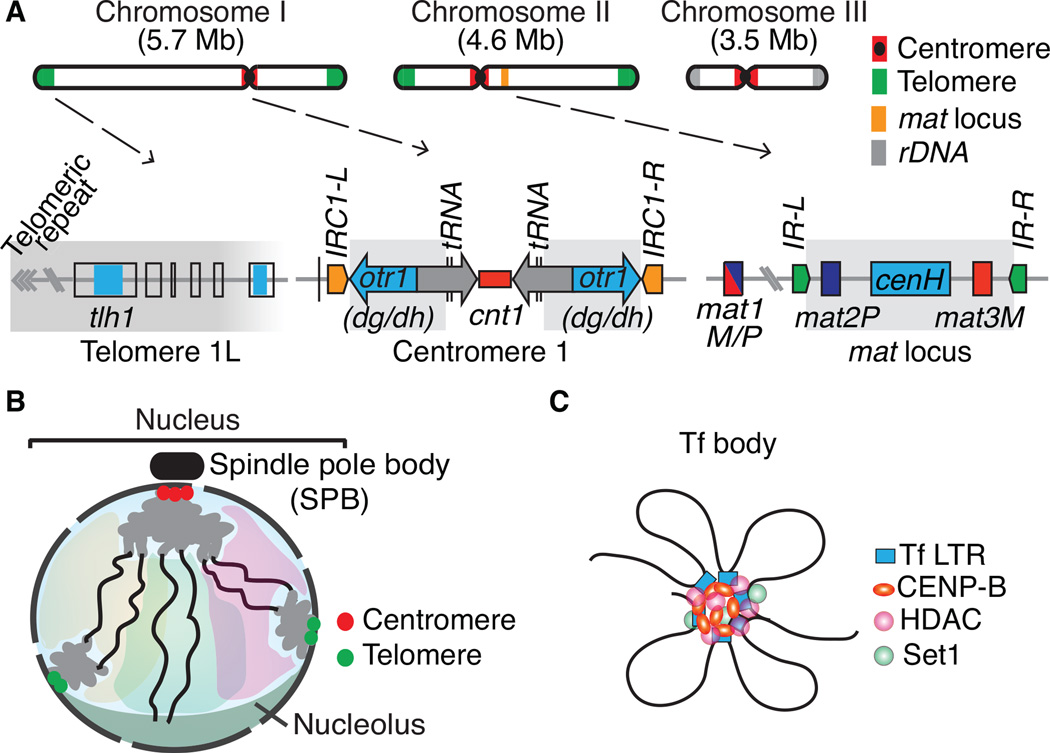
Figure 1.
Constitutive heterochromatin domains and the 3D organization of the S. pombe genome. (A) The three S. pombe chromosomes contain large blocks of heterochromatin that coats centromeres, telomeres and the silent mating-type (mat) interval. At centromeres, outer (otr) and innermost (imr) repeats surround the central core (cnt) domain, which is the site of kinetochore formation. The otr regions contain dg and dh repeats that are targets of heterochromatin formation by RNAi. tRNAs or IRC inverted repeats serve as heterochromatin boundary elements. A broad distribution of heterochromatin is also observed at the subtelomeric regions containing tlh1 and its paralogs, which contain a dh-like element within the coding region. The heterochromatin domain at the mat region contains silent mat2 and mat3 loci, which serve as donors of genetic information for the active mat1 locus. The cenH element with homology to dg and dh repeats nucleates heterochromatin, which in turn spreads across the domain surrounded by IR-L and IR-R inverted repeat boundary elements. Heterochromatin domains are highlighted in gray. (B). During interphase, chromosomes are arranged in a Rabl configuration. Interphase chromatin is subjected to various constraints and is confined to a limited sub-nuclear space (a degree of chromosome territory). (C) Tf2 retrotransposons dispersed across the genome are organized into discrete nuclear foci, called Tf bodies. CENP-B proteins collaborate with histone modifying activities such as HDACs and Set1 to form 2–3 Tf bodies in the nucleus.
In this review we summarize the findings from the fission yeast model system that have advanced our understanding of 3D genome architecture. Some reflect similar findings in higher organisms, indicating universal and fundamental genome organization principles, while others have revealed new insights and uncovered important key concepts underlying genome architecture that are also likely to universally apply.
2. Global organization of the S. pombe interphase genome
Eukaryotic chromosomes are specifically organized during interphase. S. pombe chromosomes display a polarized arrangement, in which centromeres of all three chromosomes are clustered adjacent to the spindle pole body (SPB), which is the centrosome equivalent in yeast, while telomeres are also associated with each other at the opposing hemisphere near the nuclear periphery [28] (Figure 1B). Ribosomal DNA (rDNA) repeats at the ends of chromosome III are compartmentalized within the nucleolus [29]. This polarized array is known as the Rabl configuration, which was first described in salamander larvae cells in 1885 [30]. When first observed, this configuration was thought to be a passive continuation of the chromosome configuration from the prior anaphase, in which chromosomes are pulled into the daughter cells with centromeres leading and telomeres trailing behind. However, Rabl-like configuration can be established de novo (i.e. without a prior anaphase) in both budding and fission yeast [31,32], suggesting that the polarized array of yeast chromosomes is likely not just a relic of anaphase.
Indeed, the Rabl configuration may be important for proper functioning of the genome during interphase. Sustained by both chromosome-chromosome (clustering of centromeres and telomeres) and chromosome-nuclear envelope interactions, the constraints generated by these interactions ensure that specific chromosomal regions (and genes) are confined to distinct molecular environments within the nuclear space. The positional guidance provided by the Rabl configuration may promote genome compartmentalization, which may impact the transcription of genes and the establishment of chromosome territoriality (see below). Moreover, recent evidence suggests that the interphase clustering of centromeres could provide an organizational framework to allow efficient kinetochore capture during mitosis [33]. Indeed, centromere de-clustering has been shown to correlate with defects in chromosome segregation [33]. In mammals, chromosomes surround the spindle in a ring in mitosis and meiosis, which might efficiently expose all of the kinetochores to the spindle and facilitate their capture [34,35]. Thus, leveraging the 3D organization to facilitate the intricate process of accurate and timely chromosome segregation may be a common theme among different organisms and cell types. In this regard, Rabl may reflect a purposeful and functional arrangement of the genome.
2.1 Telomere positioning requires conserved telomere-binding proteins
The polarized configuration of chromosomes in interphase S. pombe cells remains relatively fixed over time. The nuclear envelope (NE) likely provides a solid platform for anchoring centromeres and telomeres, which can limit chromosome movement and allow the Rabl arrangement to be maintained (Figure 1B). Telomeres are anchored to the NE via interactions between telomere binding proteins, such as Rap1, and the inner nuclear membrane proteins Bqt3 and Bqt4 [36]. The telomere associated protein Rap1 is recruited by the DNA binding protein Taz1, which is the human TRF ortholog. Rap1 interacts with Bqt4, and loss of Bqt4 causes the release of telomeres from the nuclear membrane in mitotic interphase, although they still reside near the nuclear periphery [36]. Since S. pombe undergoes a “closed mitosis”, in which the NE does not disintegrate, telomeres must be transiently dissociated from the NE during mitosis to facilitate proper segregation of chromosomes. This process involves phosphorylation of Rap1 by Cdc2 (the fission yeast Cyclin-dependent kinase 1), which disrupts the interaction between Rap1 and Bqt4 to induce the release of telomeres from the NE [37].
Anchoring telomeres to the nuclear periphery also requires other factors including LEM (LAP2, emerin, MAN1) domain-containing proteins, Lem2 and Man1, although the molecular mechanism remains unknown [38]. These proteins are evolutionarily related to lamina-associated polypeptide (LAP). S. pombe lacks the nuclear lamina that provides the structural framework to the NE. The inner nuclear membrane proteins Bqt3, Bqt4, Lem2 and Man1 might cooperate to maintain nuclear organization. Interestingly, the telomere bound Ku70/80 complex also participates in telomere tethering to the nuclear periphery [39]. It is therefore possible that redundant pathways maintain telomere positioning at the NE.
2.2 Centromere clustering by SUN and KASH domain proteins
Centromeres are also anchored to the periphery where they form a cluster near the SPB during interphase (Figure 1B). Two functionally distinct domains of the centromere region, the central core (cnt) and pericentromeric repeats, are spatially separated in the interphase nucleus. Whereas pericentromeric heterochromatin is located at the periphery of the clustered centromeres, the central domain protrudes toward the SPB [40]. This protrusion provides the physical docking site for the SPB. Clustering of centromeres requires factors involved in kinetochore assembly, but is independent of pericentromeric heterochromatin or microtubules [41].
Interestingly, fission and budding yeast employ distinct strategies for clustering centromeres. In S. pombe, intra-nuclear microtubules connecting the SPB and kinetochores are not apparent in interphase, and electron microscopy studies have shown that the SPB resides in the cytoplasm during interphase and only moves into the NE at the beginning of mitosis [42,43]. Studies have shown that two proteins, Sad1 (a SUN-domain protein) and Kms1 (a KASH domain containing protein), form a complex that spans the NE. Together with Csi1, a Sad1-interacting protein, this complex appears to provide the physical connection between the SPB and kinetochores [31,33,44,45]. On the other hand, in the budding yeast S. cerevisiae the SPB remains inserted within the NE throughout the cell cycle, and microtubules emanating from the SPB connect to the kinetochores to maintain centromere clustering [32,46,47].
While our understanding of Rab1 will surely continue to evolve, it is abundantly clear that key genetic elements such as centromeres and telomeres and their associating proteins not only provide nuclear landmarks, but are key determinants of configuration that also regulate chromosome dynamics (see below).
3. DNA elements involved in functional genome organization
In addition to the Rabl configuration, specific DNA elements dispersed across chromosomes are believed to contribute to the proper organization of the genome. Similar to higher eukaryotes, the S. pombe genome contains a variety of repetitive DNA elements that are organized into specialized structures. These include retrotransposons and their remnants as well as middle repetitive sequences such as tRNA genes that are dispersed across the genome. Distinct trans-acting factors associate with these loci to direct their spatial organization. Studies addressing the assembly of these DNA elements into discrete structures have broad implications for understanding the mechanisms that contribute to the functional organization of eukaryotic genomes.
3.1 Boundary elements partition the genome into distinct domains
A comprehensive analysis of the S. pombe genome identified “boundary elements” that partition heterochromatin domains at centromeres and the mat locus from the surrounding euchromatin domains [21]. Boundary elements block the spread of heterochromatin into neighboring regions [48–50]. The borders of the heterochromatin domain at the mat locus are marked by inverted repeats (IR-L and IR-R) [26,51] (Figure 1A). An abrupt transition of H3K9me and other heterochromatin factors coincides with the locations of the IR elements [52]. Heterochromatin factors also decrease at the borders of pericentromeric heterochromatin domains. Although these borders are often demarcated by clusters of tRNA genes [21,53,54], tRNA genes are absent on the right side of centromere 1 (cen1) and only a single tRNA gene is present on the left side. Instead, inverted repeat elements have been found to flank the left and the right sides of cen1 (IRC1-L/R) and cen3 (IRC3-L/R), which serve as heterochromatin barriers [53] (Figure 1A).
Multiple mechanisms are believed to contribute to boundary function. Factors recruited to boundary elements might preclude nucleosome assembly and create a chromatin environment less favorable to the spread of heterochromatin [50]. Indeed, IRC elements show a preferential enrichment for the anti-silencing factor Epe1, which promotes nucleosome turnover [55–57]. Nucleosomes are also depleted at the IR boundaries of the mat locus and tRNA loci [58,59]. The core sequence responsible for conferring boundary activity to IRs consists of multiple B-boxes, which are the binding sites for the TFIIIC transcription factor that normally initiates assembly of the RNA polymerase III (Pol III) transcription complex at tRNA and 5S RNA loci [60,61]. Interestingly, high concentrations of TFIIIC, but not Pol III, bind to the IRs in a B-box-dependent manner. Moreover, it has been shown that B-boxes are critical for boundary activity [53]. Epe1 and TFIIIC binding sites act in parallel pathways to prevent the inappropriate spread of heterochromatin that can cause deleterious gene suppression [62]. In addition to local chromatin features, it has been proposed that TFIIIC-associated sites tethered to the nuclear periphery might organize the genome and serve to partition heterochromatin from euchromatin [53].
3.2 Chromosome organizing clamps and genome organization
In addition to boundary elements, TFIIIC also localizes to a number of sites scattered across the genome without recruiting Pol III [53]. These loci, referred to as chromosome organizing clamps (COCs), are tethered to the nuclear periphery, in a manner dependent upon the B-boxes [53]. Another intriguing observation is that despite TFIIIC enrichment at all tRNA, 5S rRNA and COC sites throughout the genome, only five to ten TFIIIC foci are observed at the nuclear periphery and perinucleolar space, suggesting specific clustered interactions between TFIIIC bound loci [53]. Although the biological significance remains unknown, it is notable that a component similar to IR boundary elements may have a role in nuclear organization at COC sites. The interactions amongst COC and other TFIIIC bound loci may facilitate higher-order organization of the genome into distinct structures, perhaps loops, which are likely to impact diverse chromosomal processes. In addition to creating heterochromatin barriers, the tethering of COC sites to the nuclear periphery might also have consequences for other chromosomal processes. For example, the passage of the replication fork may be affected by the tethering of boundaries or COC sites to the nuclear periphery. Thus, tethered COC sites could serve to divide chromosomal regions into independently replicating domains. Indeed, tRNAs act as DNA replication fork pause sites [63–65]. TFIIIC and its associated factors may represent a general mechanism for partitioning the genome into distinct domains. COC-like loci, which show TFIIIC binding independent of Pol III, have been described in other species [66–69], and pioneering work by Kamakaka and colleagues has uncovered conserved functions of tRNAs in boundary function [70,71].
3.3 Transposable elements are organized into Tf bodies
Transposable elements (TE) constitute a substantial fraction of eukaryotic genomes and pose a major threat to genome stability. As a result, host genomes have evolved defense mechanisms that are critical for silencing and immobilization of transposable elements [72–74]. S. pombe contains 13 copies of the full length Tf2 retrotransposon, approximately 300 solo long terminal repeats (LTRs) and WTF repeats (often associated with TF LTRs) dispersed across the genome [75]. In addition to mechanisms such as RNA interference and other RNA processing factors that play an important role in silencing of retrotransposons [76], cells also utilize a genome surveillance mechanism that relies on conserved CENP-B proteins (Abp1, Cbh1, and Cbh2) [72]. CENP-Bs are derived from the transposases of POGO family transposons [77]. Through recruitment of chromatin-modifying activities such as histone deacetylases to assemble “closed” chromatin, CENP-Bs repress transcription of repetitive elements and render them recombinationally inert [72]. CENP-Bs also gather retrotransposons into clusters, called Tf bodies [72] (Figure 1C). The formation of clusters containing retrotransposons and their remnants may facilitate their surveillance and silencing, but clusters likely also facilitate genome organization. For example, in Drosophila, gypsy retrotransposons are clustered into specialized bodies, which are an important element of genome organization [78,79]. Moreover, in mammalian systems transposon-derived nuclear scaffold/matrix attachment region (MAR/SAR) sequences [80] are concentrated at the bases of chromatin loops through their association with SATB1 [81]. Similarly, the dimerization of CENP-B proteins bound to LTRs may connect Tf elements located nearby or at a greater map distance away. Alternatively, CENP-Bs might recruit additional factors that promote Tf clustering. Histone-modifying enzymes such as Set1 and HDACs are recruited to Tf2s and are implicated in Tf2 clustering [39,72,82–84]. However, these histone-modifying activities also target other parts of the genome. Global changes in interphase chromatin fibers may also influence clustering [84]. Collectively, these studies highlight an emerging theme in which mechanisms involved in silencing of transposons have been co-opted to control other chromosomal functions, including higher-order organization of the genome.
4. Higher order chromatin organization of the S. pombe genome
In addition to microscopy techniques that have provided critical information about the spatial organization and dynamics of specific loci, the 3C methodology and its derivatives are providing new insights into global genome organization. In general, 3C-based methods determine molecular, rather than cytological, proximity of genomic sequences and yield contact probability maps from which aspects of chromosome architecture are inferred [85]. The application of this methodology has revealed evolutionarily conserved as well as species-specific features of genome organization [13–16,86–96]. The evidence suggests that complex eukaryotic genomes are confined within chromosome territories as previously shown by cytological methods [97], and are characterized by a hierarchical organization of chromatin loops that connect genes and enhancers as well as open and closed compartments [98]. At the sub-compartment level in mammalian systems, chromatin is organized into self-interacting TAD domains, which range in size from hundreds of kilobases to several megabases, with boundaries marked by specific proteins such as the insulator binding protein CTCF and the cohesin complex as well as genomic features including tRNAs and retrotransposons [14,15,99]. Enrichment of CTCF and cohesin at the borders could imply a role in establishing and defining a TAD, but this has not yet been thoroughly addressed [100–102]. TAD-like chromosomal domains are likely a conserved principle of genome organization, although not universal [86–90,92,96]. Interestingly, TADs are further sub-divided into smaller self-interacting domains (sub-TAD). In the murine model system, specific interactions within these domains define lineage specific landscapes and are reorganized during differentiation [99].
3C-based methods have been used to probe the organization of both budding and fission yeast genomes. Evidence suggests that in the budding yeast S. cerevisiae, the Rab1 configuration serves as a dominant factor in genome organization without any detectable TAD-like domains [86,103,104]. In contrast, recent analyses of wild type and mutant S. pombe cells suggest that both the Rabl configuration and chromatin compartmentalization are fundamental elements of genome organization in fission yeast [89]. These features are discussed below.
4.1 Territorial organization of chromosomes
Chromosomal DNA is tightly condensed during mitosis in order to facilitate segregation and faithful transfer of chromosomes to daughter nuclei. However, chromatin is de-condensed during interphase to allow regulation of appropriate gene expression programs and other DNA transactions. Considering that the majority of the S. pombe genome is assembled into an open euchromatin configuration [21], it can be imagined that a chromosome might lose its territoriality after mitotic exit as de-condensation progresses with time. However, despite the level of unpacking required, early genetic and microscopy experiments in S. pombe indicated that all three chromosomes are confined to individual territories (Figure 1B) [105,106]. This feature has been explored using genome-wide chromatin interaction maps generated by Hi-C or other techniques such as Enrichment of Ligation Products (ELP) and Genome Conformation Capture (GCC) [87,89,107]. These methods all rely on the same basic principle (3C-seq method) for capturing chromatin-chromatin interactions: proximity based ligation followed by Next Generation Sequencing (NGS) [108]. Hi-C and ELP methods involve additional steps designed to enrich for the ligation junctions [85,87]. Importantly, all of these methods have confirmed the territorial feature of genome organization.
Interestingly, computational simulations, modeling and microscopy studies in other systems indicate that most genome organization features, including chromosome territoriality, naturally emerge when just the generic polymer nature of the chromatin fiber, along with a few geometric constraints, are considered [103,104,109–113]. Factors considered in the studies included the number and size of chromosomes, confinement to the nuclear space, centromere/telomere clustering, as well as the excluded volume effect from the presence of other chromosomes and the nucleolus. While these studies have provided a basic framework, there is undoubtedly more to the structural regulation of the genome. Indeed, studies suggest the existence of mechanisms that constrain chromosomes and counteract their de-condensation to promote spatial restriction and impose territoriality [89].
4.2 Specific intra- and inter-chromosomal interactions
In addition to the clustering of all three centromeres and telomeres of chromosome I and II, several specific interactions are evident in the S. pombe genome. In particular, the high-resolution Hi-C contact maps revealed a noticeable enrichment of interactions between centromere-proximal chromosomal arm regions in both intra- and inter-chromosomal arm pairs, evident by prominent cross-like patterns [89] (Figure 2A and 2B). This feature can be understood in the context of a Rab1 configuration, in which frequent interactions occur between centromere-proximal arm regions extending from centromeres that are clustered near the SPB (Figure 1B and Figure 2B). Such a cross-like pattern has been previously observed in the contact maps of S. cerevisiae, but it is believed to result from the centromeric regions of all 16 chromosomes occupying a small confined nuclear space around the SPB [86,104]. Such a model would not predict the cross-like pattern of interactions in S. pombe, which contains only three chromosomes. Thus, the unexpected observation of prominent cross-like patterns suggests that a specific mechanism promotes co-linear extension of chromosomal arms from centromeres and reinforces the Rabl conformation to promote proper genome organization.
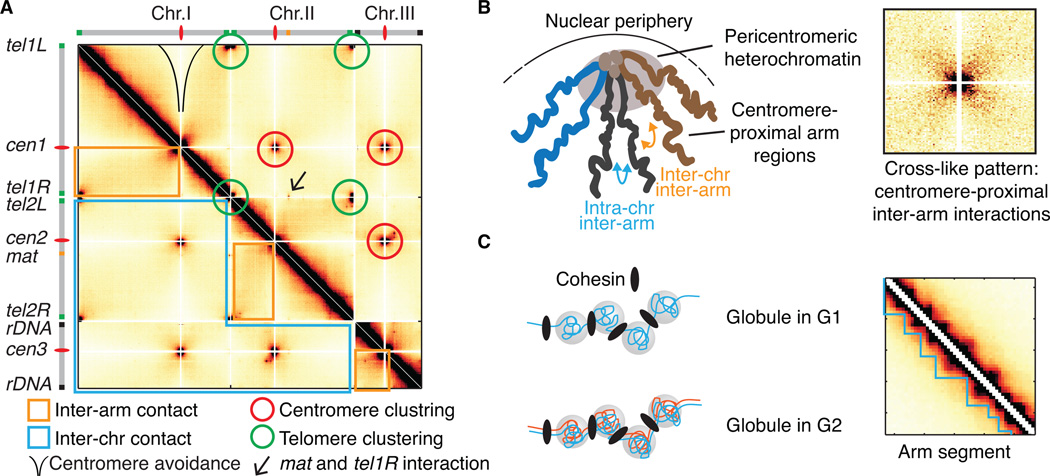
Figure 2.
General principles of S. pombe 3D genome organization as revealed by Hi-C. (A) The all-by-all contact map reveals that all three centromeres and telomeres of chromosome I and II form clusters. Centromeres avoid interactions with chromosomal arms. The specific inter-chromosomal interaction between the mat locus and tel1R suggests chromosome looping. Greater inter-arm contact compared to inter-chromosomal contact suggests chromosomal territoriality in the interphase nucleus. (B and C) Centromere-proximal inter-arm interactions and globules represent two key elements of S. pombe genome organization. The compaction of large heterochromatic domains around clustered centromeres promotes centromere-proximal intra- and inter-chromosomal inter-arm interactions and produces a cross-like interaction pattern. On chromosome arms, chromatin is organized into locally crumpled 50–100 kb repeating regions, referred to as “globules”. Globules are self-interacting domains that are observed along the diagonal of the Hi-C heatmap in both G1 and G2 cells.
Interestingly, the Hi-C study also revealed that heterochromatin-coated centromeres and telomeres avoid interaction with chromosome arms (Figure 2A). This avoidance of contact is consistent with spatial sequestration of these loci. These analyses also revealed a specific inter-chromosomal interaction between the right telomere of chromosome I (tel1R) and the mat locus on chromosome II. Consistent with tethering of a loop of chromatin to the nuclear periphery, the intra-chromosomal interaction pattern is altered at a chromosomal location coinciding with the mat locus. Tethering of specific regions to subnuclear structures is believed to be important for partitioning the chromatin into higher order loop domains [79,114,115]; thus, the mat-tel interaction may facilitate a specific chromosomal organization. However, it is important to note that chromosomal looping of the mat locus is not as frequent as centromere/telomere clustering. The mat region has also been reported to position near the SPB (and thus the centromeres) in a Clr4 dependent manner [116]. Further efforts are needed to explore how these specific interactions are formed and to understand their possible connections to biological processes such as mating-type switching, heterochromatin boundary formation and genome organization.
4.3 Globules are a prominent feature of chromosome arm architecture
Perhaps most intriguing is that interactions along the diagonal of the S. pombe Hi-C contact maps are not homogeneous. The pattern reveals the presence of local self-interacting domains ranging from 50–100 kb in size, referred to as globules [89] (Figure 2C). Globules are distributed on all three chromosomes and globule boundaries correspond to regions containing a high density of convergent genes that are enriched for cohesin [117–119]. Cohesin consists of four core subunits. The Smc1-Smc3 (structural maintenance of chromosome) heterodimer associates with the non-SMC subunits Rad21 (Mcd1 or SCC1 in other systems) and Psc3 (SA or SCC3) to form a ring-like structure that is thought to ensure cohesion by topological embrace of sister chromatids [120–123]. In addition, a role for cohesin in interphase genome organization has been described. In higher eukaryotes, cohesin cooperates with CTCF and Mediator to form loops that insulate domains and define cis-regulatory networks [124–131]. These insights are beginning to provide clues to how expression patterns unique to an organism, developmental stage or cell type can be achieved from chromatin organizational principles, and will impact our understanding of various developmental disorders [132].
Interestingly, a mutation in Rad21 disrupts globule formation in S. pombe, suggesting that cohesin is essential for their assembly. Remarkably, cohesin-dependent globules can be detected in G1 phase cells, defining a function of cohesin that is distinct from its role in sister chromatid cohesion. Consistent with this, a substantial fraction of chromatin-bound cohesin can be detected in G1 cells [89,133]. Furthermore, only a small fraction of Rad21 is cleaved and subsequently degraded during the metaphase-anaphase transition [119,120]. It is tempting to speculate that a leftover fraction of cohesin not required for sister chromatid cohesion might contribute to globule formation in Gl cells. The dynamic and unstable binding of cohesin might explain weaker globule boundaries in Gl cells as compared to those in G2 cells [120,133–136].
Exactly how cohesin contributes to globule boundary function remains unknown. The genome-wide inverse correlation between the cohesin enrichment and the relative contact probability, i.e. the more cohesin binding is observed, the more insulated the regions are on either side [89], suggests that both the position and amount of cohesin contribute to globule boundary function. This raises several possibilities. First, cohesin itself may have an architectural role in interphase chromosome organization. Cohesin-cohesin association or the ring-like structures of cohesin may form the base of loops that create highly self-interacting domains. Second, cohesin could act as cis determinants of globule boundaries either by recruiting other factors or by preventing local chromatin compaction factor(s) from spreading along the chromatin fiber. Finally, in the region between locally enriched cohesin molecules, the chromatin fiber itself may have an inherent nature for crumpling. Further work will be needed to clarify the precise role of cohesin in globule architecture and function.
These findings have provided a critical missing dimension to the role of cohesin in genome organization. The globule is the smallest chromatin organization unit yet identified, and may represent a fundamental repeating unit in chromosome arm architecture. Cohesin confines interactions within individual globule domains, and prevents interactions across the globule boundaries. Furthermore, globules comprised of crumpled chromatin may also effectively constrain chromosomes and promote their spatial organization. Indeed, defects in globules are linked to loss of chromosome territoriality [89]. Another exciting finding is that the characteristic cohesin enrichment around convergent genes of the S. pombe genome prevents inappropriate RNAPII activity [89,118]. This suggests that globules are also critical for functional genome annotation.
4.4 Heterochromatin-mediated structural constraint
Hi-C in S. pombe also revealed another fundamental organizing principle - the role of heterochromatin in imposing structural constraints on chromosomes. As mentioned above, heterochromatin factors coat large chromosomal domains at pericentromeric repeats and subtelomeric regions. The cross-like pattern of centromere-proximal interactions that are observed in wild type cells are dramatically diminished in a Clr4/Suv39h (the sole enzyme involved in methylation of H3K9 in S. pombe) mutant strain defective in heterochromatin assembly [89]. Since heterochromatin has been implicated in the preferential loading of cohesin across heterochromatin domains, the observed change could be linked to defects in cohesin localization [137,138]. However, a cohesin mutant shows no major impact on inter-arm contacts in the centromere proximal regions. Rather, heterochromatin itself may impose structural constraints to promote these interactions. Heterochromatin-mediated constraints broadly impact chromosome behavior and affect contact frequencies across chromosomal arms, supporting the predominant contribution of Rab1 configuration to genome organization. In particular, defective heterochromatin is linked to loss of chromosome territoriality and elevated levels of inter-arm and inter-chromosomal interactions. Furthermore, in cells lacking heterochromatin, regions such as centromeres, telomeres and the mat locus are less refractory to interactions with chromosomal arms [89], and the mat locus shows changes in subnuclear localization [116] that could be explained by the lessening of constraints and reduced chromosome territoriality [89].
The molecular basis for the constraints imposed by heterochromatin remains to be fully explored. However, recent evidence suggests that heterochromatin at pericentromeric regions causes compaction of the chromatin fiber that is critical for the cross-like pattern and the co-linear extension of chromosome arms from the centromeres [89]. In addition, this process may involve heterochromatin proteins such as Swi6/HPl, which has been shown to form oligomers and might bridge chromosomal arms, or alternatively, heterochromatin may serve as a recruiting platform for factors that promote interactions among heterologous chromosome regions [20,139–142]. Regardless of the mechanism, heterochromatin is expected to have a widespread and conserved role in the organization of eukaryotic genomes. Indeed, in the plant Arabidopsis thaliana, a pericentromeric cross-like interaction pattern has been described, which is consistent with the well-known organizational feature of heterochromatin-coated Arabidopsis chromocenters [143,144]. Moreover, heterochromatin islands dispersed throughout euchromatic arm regions form highly interacting nuclear structures, referred to as interactive heterochromatic islands (IHIs) or KNOT ENGAGED ELEMENT (KEE) [88,92]. In Drosophila, heterochromatin mediates the paired association that is required for achiasmate homolog disjunction in female meiosis, as well as long-range interactions at the brown locus during mitotic interphase [142,145,146].
Heterochromatin and cohesin dependent globules can be thought of as two organizational layers that likely fulfill complementary roles in the hierarchy of genome organization. Heterochromatin reinforces the Rab1 configuration by promoting strong interactions and aligning chromosomal arms to facilitate proper genome architecture, and heterochromatin coating centromeres and telomeres is believed to cause compaction at both ends of chromosomal arms to constrain the chromatin fiber. Combined with the organization of the intervening arm regions into globules, these mechanisms likely promote chromosome restriction within the three dimensional nuclear space. Indeed, either the loss of cohesin required for the proper assembly of globules or the loss of heterochromatin assembly causes an increase in inter-chromosomal contacts and a reduction in chromosomal territoriality [89].
4.5 Functional sub-nuclear environments in interphase cells
The organization of the interphase nucleus can create specific nuclear microenvironments for various nuclear processes, such as transcription, replication and DNA repair. The ELP method in S. pombe revealed a statistically significant spatial proximity among highly expressed genes, suggesting functionally active foci are efficiently transcribed in close association [87]. This is indicative of a sub-nuclear environment similar to the described mammalian transcription factory [147]. Genes expressed in G2 phase and 23 gene ontology groups also showed significant spatial proximity. Interestingly, several shared DNA motifs within the promoter regions of these genes suggest that DNA binding proteins may mediate recruitment to a functional nuclear environment. For example, the artificial substitution of the IR-R boundary with rDNA repeats directs the mat region to the peri-nucleolar space. The sequence-specific DNA binding protein Reb1, which binds to replication pausing sites (Ter) in the rDNAs [148], mediates this re-positioning. Ter sites present at chromosomal locations also make contact through Reb1 dimerization and cooperatively modulate replication termination [149].
More recently, the GCC method, polymer modeling and DNA adenine methyltransferase identification (DamID) have been used to map the binding sites of nuclear membrane proteins and have provided further evidence of functional sub-nuclear environments that correlate with gene expression activity [107,150]. The radial re-positioning of chromatin relative to the NE correlates with transcriptional change in various systems, although whether this is cause or consequence remains to be determined. In S. pombe, these studies suggest more internal locations of actively expressed genes, and nuclear peripheral localization of poorly expressed genes [107,150]. Intriguingly, chromosome regions containing gene clusters that are up-regulated upon nitrogen starvation re-position from the nuclear periphery to the interior with accompanying transcriptional up-regulation in response to nitrogen depletion. This nuclear re-organization is regulated by the class II histone deacetylase Clr3, which represses these loci [151].
5. Dynamic regulation of the S. pombe genome
It has been shown that particular genes or chromosome regions exhibit a degree of mobility in the interphase nucleus, which causes cell-to-cell variability in nuclear positioning [112,113,152,153]. In addition to such local chromatin mobility in mitotically dividing cells, large-scale movement/repositioning of chromosomes has also been observed following entry into meiosis in S. pombe. In this section, we discuss the highly dynamic nature of the chromatin fiber and chromosome movements.
5.1 A role for condensin in genome organization in mitotic cells
Chromosomal DNA that is loosely packed in the interphase nucleus is dramatically reorganized into a more condensed form of chromatin during mitosis. Condensin functions as a major effector for condensation, and various aspects of its activity are regulated during the cell cycle [154]. Phosphorylation of the SMC subunit Cut3 (SMC4 in other systems) during entry into mitosis induces nuclear accumulation of condensin [155], although small amounts of condensin are detected even on interphase chromosomes [119,156,157]. Condensin shows preferential enrichment at the centromere central core and the rDNAs, as well as at Pol III transcribed genes such as tRNA and 5S rRNA on chromosome arms [39,156,158–160]. Exactly how condensin achieves chromosome condensation is still under investigation, but it is plausible that condensin-bound loci along the chromosome are brought together by topological embrace or multimerization of condensin [161].
Recent evidence indicates that condensin impacts genome organization by controlling the spatial positioning of tRNA and 5S rRNA near the centromeres. Pol III transcription machinery is believed to recruit condensin to these loci, thereby physically linking them to condensin-enriched centromeres [156,162] (Figure 3). Condensin also affects clustering of Tf2 retrotransposons near centromeres [39]. CENP-B proteins and the Ku heterodimer recruit condensin, and these factors collaborate to cluster Tf2 retrotransposons [39] (Figure 3). Intriguingly, Tf2 organization exhibits dynamic behavior through the cell cycle. Acetylation of histone H3 lysine 56 affects condensin localization at Tf2, and modulates the integrity and centromeric proximity of Tf2 bodies during S phase and upon DNA damage [39].
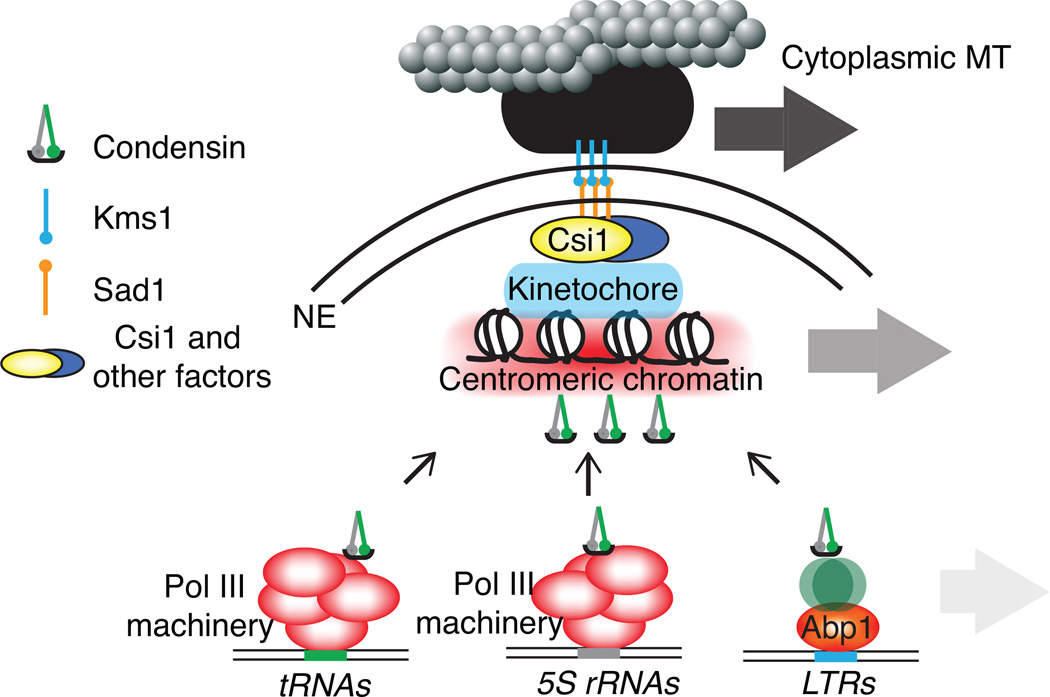
Figure 3.
Dynamic organization of interphase chromatin. Condensin regulates the spatial proximity of dispersed genetic elements such as tRNAs, 5S rRNAs and LTRs to the centromere. Condensin is recruited through distinct protein complexes. Condensin-mediated associations may facilitate chromosomal movements driven by microtubule dependent oscillation of the SPB. A molecular bridge formed by Csil and presumably other factors transmits the directional movement of the SPB to centromeres and condensin associated loci (large arrow).
Condensin-mediated organization is believed to contribute to interphase chromosome movement. Cytoplasmic microtubules cause SPB oscillation along the longitudinal axis of the cell [163]. This movement correlates with directional movement of centromeres, which are attached to the SPB by specific connector molecules [153]. However, a surprising finding is that the centromeric motion can be transmitted to genetic elements, such as Pol III transcribed loci and LTRs, which are clustered near the centromere through condensin [153](Figure 3). Thus, these molecular connections between chromosomes and cytoskeleton components establish much more than just a static association. In fact, they provide the controlling forces behind the interphase configuration. Further work is needed to understand the biological significance of these findings.
5.2 Genome re-organization upon meiotic induction
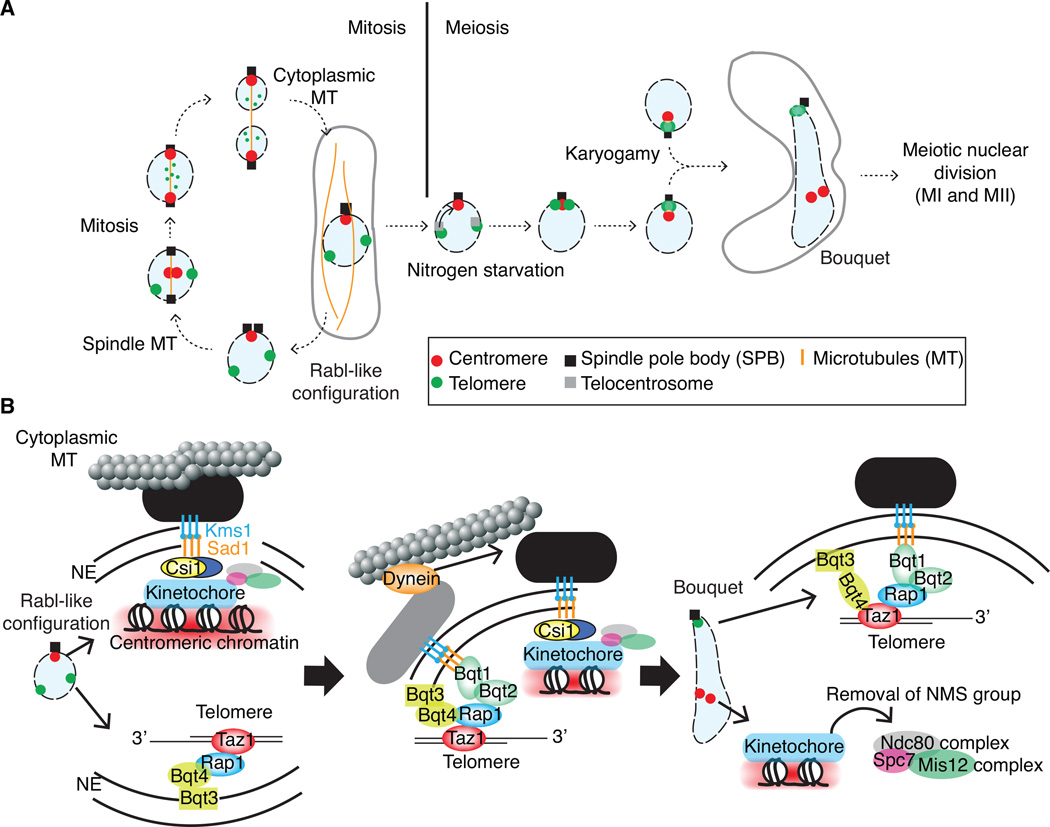
A dynamic re-organization of the genome occurs in response to environmental or developmental stimuli. One striking example occurs upon entry into meiosis. S. pombe proliferates as a haploid organism in ideal, nutrient-rich environments. However, upon nitrogen starvation, cells of opposite mating-type conjugate to form a zygote containing a diploid nucleus, and subsequently enter meiosis to produce four haploid spores. Dramatic changes in genome organization are observed as cells undergo meiosis (Figure 4A). The Rab1 configuration of interphase cells changes to a bouquet configuration, in which telomeres and centromeres undergo positional switching in relation to the SPB. During this reorganization, telomeres relocate from their distal tethered positions to cluster together near the SPB, displacing clustered centromeres [164]. Studies of bouquet defective mutants (e.g. cells lacking the telomere binding Taz1 protein or the KASH domain protein Kms1 that is important for meiotic SPB integrity) indicate that the proper structure and/or function of both telomeres and the SPB are required for bouquet formation [165–168]. These results suggest that the nature of local anchoring devices may drive the striking reorganization of the genome during meiosis.
Figure 4.
Dynamic reconfiguration of chromosomes occurs during mitosis and meiosis. (A) The Rabl-like configuration in the interphase nucleus is transiently perturbed during mitosis. Centromeres dissociate from the SPB, while telomeres are de-clustered and released from the NE, liberating chromosomes for mitotic separation. Centromeres are recaptured by spindle microtubules, which are nucleated by the SPB buried in the NE, for proper chromosome segregation. Upon meiotic induction, the chromosomes reconfigure to form a polarized chromosomal array called the bouquet. This process is driven by dynamic cytoskeleton associated activities involving the SPB, telocentrosomes and motor proteins. The bouquet arrangement is achieved in two steps: (1) telomere movement to achieve bouquet clustering at the SPB and (2) centromere dissociation from the SPB. The functional 3D microenvironment created by these processes is critical for proper meiotic function (see text). Bundled telomeres at the SPB lead the nuclear oscillatory movements between the cell poles (horsetail shaped nucleus) for homologous pairing. After movement stops, chromosomes condense and cells initiate meiotic nuclear division. (B) Centromeres and telomeres are tethered to the NE through specific protein-protein interactions. The formation of the bouquet chromosome configuration is accomplished through changes in the molecular connections between chromatin associating proteins and nuclear membrane proteins, and is guided by dynamic cytoskeleton rearrangements.
An important question remained - how do telomeres undergo such a dramatic physical relocation in the context of 3D nuclear space? Nitrogen starvation and the pheromone response cause the up-regulation of the meiosis specific connector proteins Bqt1 and Bqt2, which interact with the Rap1-Taz1 telomere complex and bridge telomeres to the Sad1-Kms1 complex (Figure 4B). Time-lapse microscopy revealed multiple Sadl-Bqt1/2-Rapl intermediate foci, suggesting a transient dispersal of Sadl from the SPB to telomeres. These Sadl-Bqtl/2-Rapl foci that mark telomeres ultimately gather at the SPB to form the bouquet [169]. This step-wise process, which is facilitated by microtubules emanating from both the SPB and a meiosis specific microtubule-organizing center (called the “telocentrosome”), underlies the dramatic reorganization of the genome during the sexual differentiation process [164,170,171].
5.3 Dramatic chromosome reconfiguration for meiotic function
The reorganization of the genome during meiosis is linked to important biological functions. The clustering of telomeres at the SPB provides the physical basis for vigorous chromosomal movement during meiotic prophase. This process involves microtubule arrays emanating from the SPB, aided by motor activity, which drive chromosomal motion and create an elongated “horsetail” shaped nucleus [170,172,173] (Figure 4A). Cells defective in either bouquet formation or horsetail movement show abnormal pairing along the chromosome arms and reduced meiotic recombination [165–167,170,174]. In contrast, recombination between ectopic sites increases in mutants defective in horsetail movement [167]. These results strongly suggest that telomere-led movement coupled to nuclear oscillation eliminates unwanted interactions and promotes homologous pairing. In addition to chromosome alignment by horsetail nuclear movement, meiosis-specific non-coding RNAs promote local homologous pairing [175].
The formation of the bouquet is not simply a requirement for horsetail nuclear movement. Recent studies suggest an additional biological significance for the bouquet. Surprisingly, the bouquet was found to be crucial for regulating the SPB and for proper meiotic spindle function in meiotic nuclear division [176–178]. During the step-wise meiotic bouquet formation process, the SUN-KASH-cytoskeletal system temporarily places the telomeres and centromeres in close proximity to the SPB [164,179]. It has been suggested that telomere chromatin contact with the SPB likely confers the ability to promote meiotic spindle assembly [176–178]. In another striking finding, it was reported that bouquet-deficient cells show defects in the localization of kinetochore proteins and the centromeric histone H3 variant CENP-A, which in turn causes spindle attachment failure [180]. Moreover, the formation of heterochromatin at centromeres is impaired in these cells [180]. These results support a model that centromeres have a tendency to disassemble during meiosis and the dynamic reorganization of chromosomes might bring centromeres to a telomere-proximal microenvironment conducive for the proper reassembly of centromeres [180]. During meiotic prophase, dissociation of the kinetochore NMS (Ndc80-Mis12-Spc7) complex is triggered by pheromone signaling and is linked to displacement of clustered centromeres from the SPB [179,181]. The temporary dissociation of centromeres from the SPB might also provide a window of opportunity to reorganize the centromere/kinetochore in the context of bouquet configuration. Indeed, it has been suggested that the bouquet may be necessary to prepare centromeres for meiotic nuclear division, for example by loading meiosis specific factors that ensure kinetochore mono-orientation [179,181].
In sum, chromosome reorganization and nuclear movement are linked to dramatic cytoskeleton rearrangements that occur in response to environmental cues. The resultant chromosome re-configuration may enable the specialized functional response required for various meiotic events. These studies have uncovered important aspects of the molecular mechanism and function of the SUN-KASH-cytoskeletal system. These components are evolutionarily conserved and are referred to as the LINC (linker of nucleoskeleton and cytoskeleton) complex. LINC has been found to play a critical role in many biological functions that, when disrupted, can underlie disease [182–184].
6. Conclusions/Discussion
Despite rapid recent progress, our view of interphase chromatin organization is still limited. Interphase chromatin is highly dynamic while being confined to a limited subnuclear space consistent with nuclear territoriality [185]. In S. pombe, interphase chromatin is subject to both random and coordinated motion from centromere oscillation. Although seemingly contradictory, dynamic movements are not inconsistent with territorial organization. It is the dynamic motion of chromatin confined within various constraints that yields the characteristic static snapshot of chromosome architecture provided by microscopy and 3C techniques. While genome-wide 3C methods provide important information needed to understand genome organization, it is important to interpret the output from these methods as probabilistic datasets. Such a viewpoint allows the appreciation that these datasets are really ensemble and seemingly static representations of what is actually a very dynamic 3D genome organization [186]. Despite these limitations, high resolution Hi-C has uncovered important genome organizational principles and has provided surprising insights into the roles of cohesin and heterochromatin in imposing constraints on interphase chromosomes. Conserved factors including cohesin and heterochromatin proteins that are involved in genome organization are also implicated in a wide range of other biological functions, such as transcriptional regulation, chromosome segregation and DNA repair. Further work will be needed to gain a better understanding of the exact nature of the structure-function relationships among these factors. The combination of genome-wide 3C analysis with yeast genetics in S. pombe promises to provide a powerful system to dissect key mechanistic details of genome packaging.
Acknowledgements
We regret that we were unable to cite all work due to space constraints. We thank Drs. Julia Promisel Cooper, Tomoyasu Sugiyama and Hernan Diego Folco for comments. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Glossary
Abbreviations
- 3C
chromosome conformation capture
- mat
mating-type locus
- SPB
spindle pole body
- rDNA
ribosomal DNA
- NE
nuclear envelope
- LEM
LAP2, emerin, MAN1
- LAP
lamina-associated peptide
- cnt
centromere central core
- IR
inverted repeat
- cen
centromere
- Pol III
RNA Polymerase III
- COC
chromosome organizing clamps
- TE
transposable element
- LTR
long terminal repeat
- WTF
with TF LTRs
- MAR/SAR
matrix/scaffold attachment region
- ELP
enrichment of ligation products
- GCC
genome conformation capture
- KEE
knot engaged element
- IHI
interactive heterochromatic islands
- NMS
Ndc80-Mis12-Spc7
- LINC
linker of nucleoskeleton and cytoskeleton
- TAD
topologically associating domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 2.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Zimmer C, Fabre E. Principles of chromosomal organization: lessons from yeast. J. Cell Biol. 2011;192:723–733. doi: 10.1083/jcb.201010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai S, Heun P, Gasser SM. Roles for nuclear organization in the maintenance of genome stability. Epigenomics. 2010;2:289–305. doi: 10.2217/epi.09.49. [DOI] [PubMed] [Google Scholar]
- 5.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aparicio OM. Location, location, location: it’s all in the timing for replication origins. Genes Dev. 2013;27:117–128. doi: 10.1101/gad.209999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pai DA, Engelke DR. Spatial organization of genes as a component of regulated expression. Chromosoma. 2010;119:13–25. doi: 10.1007/s00412-009-0236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 9.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 11.Wurtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 2006;14:477–495. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 12.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 13.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sexton T, et al. Three-dimensional folding and functional organization principles of theDrosophila genome . Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Rapkin LM, Anchel DR, Li R, Bazett-Jones DP. A view of the chromatin landscape. Micron. 2012;43:150–158. doi: 10.1016/j.micron.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Rouquette J, Cremer C, Cremer T, Fakan S. Functional nuclear architecture studied by microscopy: present and future. Int. Rev. Cell Mol. Biol. 2010;282:1–90. doi: 10.1016/S1937-6448(10)82001-5. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa 0, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 21.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 22.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 24.Grewal SI, Klar AJ. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- 26.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 27.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 28.Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzawa S, Yanagida M. 5Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 1992;101(Pt 2):267–275. doi: 10.1242/jcs.101.2.267. [DOI] [PubMed] [Google Scholar]
- 30.Rabl C. On Cell Division. Morphologisches Jahrbuch. 1885;10:214–330. [Google Scholar]
- 31.Goto B, Okazaki K, Niwa o. Cytoplasmic microtubular system implicated in de novo formation of a Rabl-like orientation of chromosomes in fission yeast. J. Cell Sci. 2001;114:2427–2435. doi: 10.1242/jcs.114.13.2427. [DOI] [PubMed] [Google Scholar]
- 32.Jin QW, Fuchs J, Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 2000;113(Pt 11):1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- 33.Hou H, et al. Csi1 links centromeres to the nuclear envelope for centromere clustering. J. Cell Biol. 2012;199:735–744. doi: 10.1083/jcb.201208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 36.Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, Haraguchi T, Hiraoka Y. Membrane proteins Bqt3 and −4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 2009;187:413–427. doi: 10.1083/jcb.200902122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita I, et al. Telomere-nuclear envelope dissociation promoted by Rap1 phosphorylation ensures faithful chromosome segregation. Curr. Biol. 2012;22:1932–1937. doi: 10.1016/j.cub.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez Y, Saito A, Sazer S. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus (Calcutta) 2012;3:60–76. doi: 10.4161/nucl.18824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka A, et al. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol. Cell. 2012;48:532–546. doi: 10.1016/j.molcel.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kniola B, O’Toole E, McIntosh JR, Mellone B, Allshire R, Mengarelli S, Hultenby K, Ekwall K. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. cell. 2001;12:2767–2775. doi: 10.1091/mbc.12.9.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Appelgren H, Kniola B, Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. doi: 10.1242/jcs.00707. [DOI] [PubMed] [Google Scholar]
- 42.Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uzawa S, Li F, Jin Y, McDonald KL, Braunfeld MB, Agard DA, Cande WZ. Spindle pole body duplication in fission yeast occurs at the G1/S boundary but maturation is blocked until exit from S by an event downstream of cdc10+ . Mol. Biol. Cell. 2004;15:5219–5230. doi: 10.1091/mbc.E04-03-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genomics. 2004;270:449–461. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- 46.Dorn JF, Jaqaman K, Rines DR, Jelson GS, Sorger PK, Danuser G. Yeast kinetochore microtubule dynamics analyzed by high-resolution three-dimensional microscopy. Biophys. J. 2005;89:2835–2854. doi: 10.1529/biophysj.104.058461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- 48.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 49.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 51.Thon G, Bjerling P, Bunner CM, Verhein-Hansen J. Expression-state boundaries in the mating-type region of fission yeast. Genetics. 2002;161:611–622. doi: 10.1093/genetics/161.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 53.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 54.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 55.Ayoub N, Noma K, Isaac S, Kahan T, Grewal SI, Cohen A. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell. Biol. 2003;23:4356–4370. doi: 10.1128/MCB.23.12.4356-4370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zofall M, Grewal SI. Swi6/HPl recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Aygun 0, Mehta S, Grewal SI. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol. 2013;20:547–554. doi: 10.1038/nsmb.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI. Asfl/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol. Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia JF, Dumesic PA, Hartley PD, El-Samad H, Madhani HD. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 2010 doi: 10.1101/gad.1946410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donze D. Extra-transcriptional functions of RNA Polymerase HI complexes: TFIIIC as a potential global chromatin bookmark. Gene. 2012;493:169–175. doi: 10.1016/j.gene.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 61.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase HI transcription control elements: themes and variations. Gene. 2012;493:185–194. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 62.Garcia JF, Al-Sady B, Madhani HD. Intrinsic Toxicity of Unchecked Heterochromatin Spread Is Suppressed by Redundant Chromatin Boundary Functions in Schizosacchromyces pombe. 2015:G3. doi: 10.1534/g3.115.018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 64.McFarlane RJ, Whitehall SK. tRNA genes in eukaiyotic genome organization and reorganization. Cell Cycle. 2009;8:3102–3106. doi: 10.4161/cc.8.19.9625. [DOI] [PubMed] [Google Scholar]
- 65.Pryce DW, Ramayah S, Jaendling A, McFarlane RJ. Recombination at DNA replication fork barriers is not universal and is differentially regulated by Swil. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4770–4775. doi: 10.1073/pnas.0807739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase HI machinery inSaccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol. Cell. Biol. 2004;24:4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase HI transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oler AJ, et al. Human RNA polymerase HI transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010;17:620–648. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase HI transcription complexes in human cells. Nat. Struct. Mol. Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers inSaccharomyces cerevisiae . EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain inSaccharomyces cerevisiae . Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 73.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ebina H, Levin HL. Stress management: how cells take control of their transposons. Mol. Cell. 2007;27:180–181. doi: 10.1016/j.molcel.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 75.Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence ofSchizosaccharomyces pombe . Genome Res. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamanaka S, Mehta S, Reyes-Turcu FE, Zhuang F, Fuchs RT, Rong Y, Robb GB, Grewal SI. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–560. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Casola C, Hucks D, Feschotte C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 79.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 80.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 81.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 82.Lorenz DR, Mikheyeva IV, Johansen P, Meyer L, Berg A, Grewal SI, Cam HP. CENP-B cooperates with Set1 in bidirectional transcriptional silencing and genome organization of retrotransposons. Mol. Cell. Biol. 2012;32:4215–4225. doi: 10.1128/MCB.00395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lorenz DR, Meyer LF, Grady PJ, Meyer MM, Cam HP. Heterochromatin assembly and transcriptome repression by Setl in coordination with a class II histone deacetylase. Elife. 2014:3. doi: 10.7554/eLife.04506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mikheyeva IV, Grady PJ, Tamburini FB, Lorenz DR, Cam HP. Multifaceted genome control by Setl Dependent and Independent of H3K4 methylation and the Set1C/COMPASS complex. PLOS Genet. 2014;10:el004740. doi: 10.1371/journal.pgen.1004740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanizawa H, Iwasaki 0, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, Jacobsen SE. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis . Mol. Cell. 2014;55:694–707. doi: 10.1016/j.molcel.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mizuguchi T, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe . Nature. 2014;516:432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Le TB, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of theDrosophila genome into physical domains. Mol. Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grob S, Schmid MW, Grossniklaus U. Hi-C analysis inArabidopsisidentifies theKNOTa structure with similarities to theflamenco locus of Drosophila . Mol. Cell. 2014;55:678–693. doi: 10.1016/j.molcel.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 93.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert JP, Noble WS, Le Roch KG. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014;24:974–988. doi: 10.1101/gr.169417.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:el57. [Google Scholar]
- 98.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol. Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBOJ. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seitan VC, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U. S. A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wong H, Marie-Nelly H, Herbert S, Carrivain P, Blanc H, Koszul R, Fabre E, Zimmer C. A predictive computational model of the dynamic 3D interphase yeast nucleus. Curr. Biol. 2012;22:1881–1890. doi: 10.1016/j.cub.2012.07.069. [DOI] [PubMed] [Google Scholar]
- 104.Tjong H, Gong K, Chen L, Alber F. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res. 2012;22:1295–1305. doi: 10.1101/gr.129437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Molnar M, Kleckner N. Examination of interchromosomal interactions in vegetatively growing diploidSchizosaccharomyces pombe cells by Cre/loxP site-specific recombination. Genetics. 2008;178:99–112. doi: 10.1534/genetics.107.082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scherthan H, Bahler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeastJ . Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grand RS, Pichugina T, Gehlen LR, Jones MB, Tsai P, Allison JR, Martienssen R, O’Sullivan JM. Chromosome conformation maps in fission yeast reveal cell cycle dependent sub nuclear structure. Nucleic Acids Res. 2014;42:12585–12599. doi: 10.1093/nar/gku965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Laat W, Dekker J. 3C-based technologies to study the shape of the genome. Methods. 2012;58:189–191. doi: 10.1016/j.ymeth.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008;4:el000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Therizols P, Duong T, Dujon B, Zimmer C, Fabre E. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2025–2030. doi: 10.1073/pnas.0914187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 2005;168:375–387. doi: 10.1083/jcb.200409091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marshall WF, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 113.Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- 114.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 115.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 116.Alfredsson-Timmins J, Henningson F, Bjerling P. The Clr4 methyltransferase determines the subnuclear localization of the mating-type region in fission yeast. J. Cell Sci. 2007;120:1935–1943. doi: 10.1242/jcs.03457. [DOI] [PubMed] [Google Scholar]
- 117.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes inS. pombe . Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 119.Schmidt CK, Brookes N, Uhlmann F. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 2009;10:R52. doi: 10.1186/gb-2009-10-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 122.Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat. Cell Biol. 2011;13:1170–1177. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 123.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality.. Annu. Rev. Cell Dev. Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 124.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 126.Degner SC, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of theIgh locus and antisense transcription in pro-B cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 128.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulatedIFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBOJ. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nativio R, et al. Cohesin is required for higher-order chromatin conformation at the imprintedIGF2-H1 9 locus. PLOS Genet. 2009;5:el000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bose T, Gerton JL. Cohesinopathies, gene expression, and chromatin organizationJ . Cell Biol. 2010;189:201–210. doi: 10.1083/jcb.200912129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bernard P, et al. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBOJ. 2008;27:111–121. doi: 10.1038/sj.emboj.7601955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr. Biol. 2006;16:875–881. doi: 10.1016/j.cub.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 135.Vaur S, Feytout A, Vazquez S, Javerzat JP. Pds5 promotes cohesin acetylation and stable cohesin-chromosome interaction. EMBO Rep. 2012;13:645–652. doi: 10.1038/embor.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Feytout A, Vaur S, Genier S, Vazquez S, Javerzat JP. Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Esol-mediated sister chromatid cohesion by antagonizing Wpl1. Mol. Cell. Biol. 2011;31:1771–1786. doi: 10.1128/MCB.01284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 138.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HPl in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 139.Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 140.Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol. Cell. Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Renauld H. Heterochromatin: a meiotic matchmaker? Trends Cell Biol. 1997;7:201–205. doi: 10.1016/S0962-8924(97)01034-9. [DOI] [PubMed] [Google Scholar]
- 143.Moissiard G, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Grob S, Schmid MW, Luedtke NW, Wicker T, Grossniklaus U. Characterization of chromosomal architecture inArabidopsis by chromosome conformation capture. Genome Biol. 2013;14:R129. doi: 10.1186/gb-2013-14-11-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr. Opin. Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 146.Wolf KW. How meiotic cells deal with non-exchange chromosomes. Bioessays. 1994;16:107–114. doi: 10.1002/bies.950160207. [DOI] [PubMed] [Google Scholar]
- 147.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat. Rev. Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 148.Jakociunas T, Domange Jordo M, Ait Mebarek M, Bunner CM, Verhein-Hansen J, Oddershede LB, Thon G. Subnuclear relocalization and silencing of a chromosomal region by an ectopic ribosomal DNA repeat. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E4465–E4473. doi: 10.1073/pnas.1315581110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Singh SK, Sabatinos S, Forsburg S, Bastia D. Regulation of replication termination by Rebl protein-mediated action at a distance. Cell. 2010;142:868–878. doi: 10.1016/j.cell.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Steglich B, Filion GJ, van Steensel B, Ekwall K. The inner nuclear membrane proteins Man1 and Ima1 link to two different types of chromatin at the nuclear periphery inS. pombe . Nucleus (Calcutta) 2012;3:77–87. doi: 10.4161/nucl.18825. [DOI] [PubMed] [Google Scholar]
- 151.Alfredsson-Timmins J, Kristell C, Henningson F, Lyckman S, Bjerling P. Reorganization of chromatin is an early response to nitrogen starvation inSchizosaccharomyces pombe . Chromosoma. 2009;118:99–112. doi: 10.1007/s00412-008-0180-6. [DOI] [PubMed] [Google Scholar]
- 152.Vazquez J, Belmont AS, Sedat JW. Multiple regimes of constrained chromosome motion are regulated in the interphaseDrosophila nucleus. Curr. Biol. 2001;11:1227–1239. doi: 10.1016/s0960-9822(01)00390-6. [DOI] [PubMed] [Google Scholar]
- 153.Kim KD, Tanizawa H, Iwasaki 0, Corcoran CJ, Capizzi JR, Hayden JE, Noma K. Centromeric motion facilitates the mobility of interphase genomic regions in fission yeast. J. Cell Sci. 2013;126:5271–5283. doi: 10.1242/jcs.133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 155.Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Iwasaki 0, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell. 2010;21:254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 2002;417:197–202. doi: 10.1038/417197a. [DOI] [PubMed] [Google Scholar]
- 158.Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K, Yanagida M. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J. Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- 161.Cuylen S, Haering CH. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 2011;21:552–559. doi: 10.1016/j.tcb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 162.Iwasaki 0, Noma K. Global genome organization mediated by RNA polymerase Ill-transcribed genes in fission yeast. Gene. 2012;493:195–200. doi: 10.1016/j.gene.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chikashige Y, Ding DQ, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeastSchizosaccharomyces pombe . EMBOJ. 1997;16:193–202. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- 166.Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants ofSchizosaccharomyces pombe . Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- 167.Niwa 0, Shimanuki M, Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBOJ. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa 0. A novel fission yeast gene, kmsl+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- 169.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqtl and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 170.Yamamoto A, West RR, Mcintosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. JCell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yoshida M, et al. Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. J. Cell Biol. 2013;200:385–395. doi: 10.1083/jcb.201207168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 173.Funaya C, et al. Transient structure associated with the spindle pole body directs meiotic microtubule reorganization inS. pombe . Curr. Biol. 2012;22:562–574. doi: 10.1016/j.cub.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 175.Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, Hiraoka Y. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science. 2012;336:732–736. doi: 10.1126/science.1219518. [DOI] [PubMed] [Google Scholar]
- 176.Tomita K, Cooper JP. The telomere bouquet controls the meiotic spindle. Cell. 2007;130:113–126. doi: 10.1016/j.cell.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 177.Tomita K, Bez C, Fennell A, Cooper JP. A single internal telomere tract ensures meiotic spindle formation. EMBO Rep. 2013;14:252–260. doi: 10.1038/embor.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fennell A, Fernandez-Alvarez A, Tomita K, Cooper JP. Telomeres and centromeres have interchangeable roles in promoting meiotic spindle formationJ . Cell Biol. 2015;208:415–428. doi: 10.1083/jcb.201409058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol. Biol. Cell. 2005;16:2325–2338. doi: 10.1091/mbc.E04-11-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Klutstein M, Fennell A, Fernandez-Alvarez A, Cooper JP. The telomere bouquet regulates meiotic centromere assembly. Nat. Cell Biol. 2015;17:458–469. doi: 10.1038/ncb3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hayashi A, Asakawa H, Haraguchi T, Hiraoka Y. Reconstruction of the kinetochore during meiosis in fission yeastSchizosaccharomyces pombe . Mol. Biol. Cell. 2006;17:5173–5184. doi: 10.1091/mbc.E06-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mejat A, Misteli T. LINC complexes in health and disease. Nucleus (Calcutta) 2010;1:40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics.. Dev. Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 184.Yamamoto A. Gathering up meiotic telomeres: a novel function of the microtubule-organizing center. Cell. Mol. Life Sci. 2014;71:2119–2134. doi: 10.1007/s00018-013-1548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chuang CH, Belmont AS. Moving chromatin within the interphase nucleus-controlled transitions? Semin. Cell Dev. Biol. 2007;18:698–706. doi: 10.1016/j.semcdb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fudenberg G, Mirny LA. Higher-order chromatin structure: bridging physics and biology. Curr. Opin. Genet. Dev. 2012;22:115–124. doi: 10.1016/j.gde.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]