Figure 1.
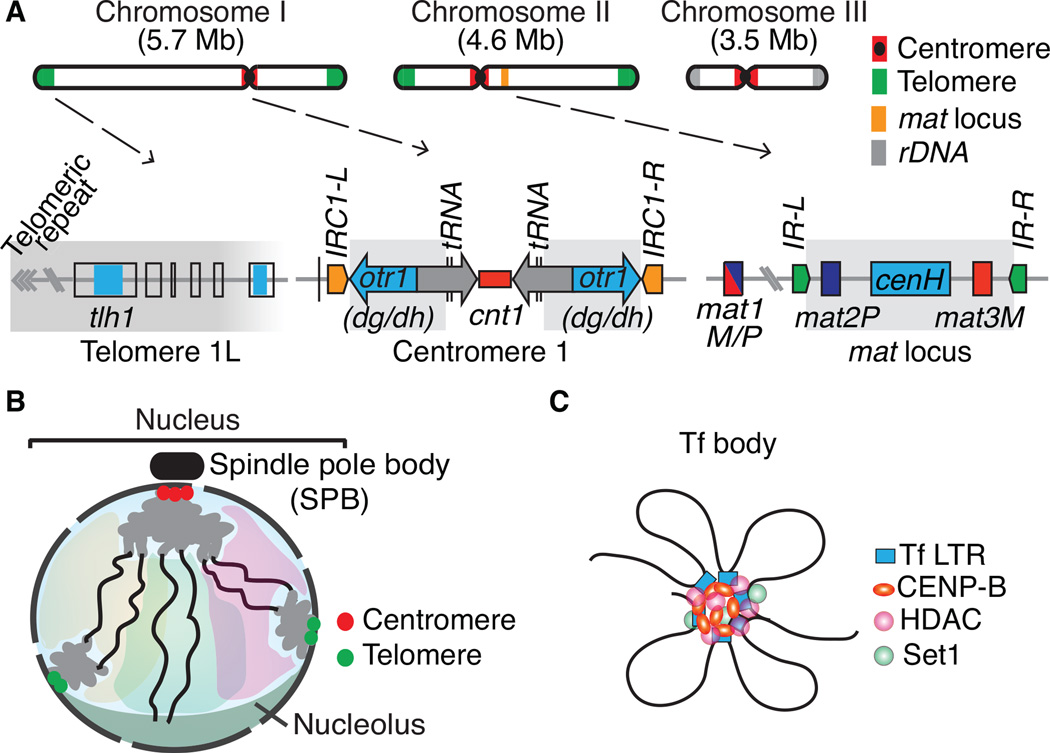
Constitutive heterochromatin domains and the 3D organization of the S. pombe genome. (A) The three S. pombe chromosomes contain large blocks of heterochromatin that coats centromeres, telomeres and the silent mating-type (mat) interval. At centromeres, outer (otr) and innermost (imr) repeats surround the central core (cnt) domain, which is the site of kinetochore formation. The otr regions contain dg and dh repeats that are targets of heterochromatin formation by RNAi. tRNAs or IRC inverted repeats serve as heterochromatin boundary elements. A broad distribution of heterochromatin is also observed at the subtelomeric regions containing tlh1 and its paralogs, which contain a dh-like element within the coding region. The heterochromatin domain at the mat region contains silent mat2 and mat3 loci, which serve as donors of genetic information for the active mat1 locus. The cenH element with homology to dg and dh repeats nucleates heterochromatin, which in turn spreads across the domain surrounded by IR-L and IR-R inverted repeat boundary elements. Heterochromatin domains are highlighted in gray. (B). During interphase, chromosomes are arranged in a Rabl configuration. Interphase chromatin is subjected to various constraints and is confined to a limited sub-nuclear space (a degree of chromosome territory). (C) Tf2 retrotransposons dispersed across the genome are organized into discrete nuclear foci, called Tf bodies. CENP-B proteins collaborate with histone modifying activities such as HDACs and Set1 to form 2–3 Tf bodies in the nucleus.