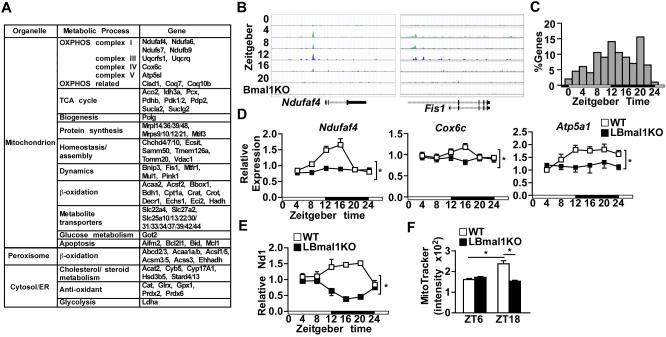
Figure 5. The oxidative metabolism pathway is a primary transcriptional target of the hepatic clock.
(A) Partial list of the 211 genes involved in mitochondrial function that are bound by Bmal1, Clock, and Cry1 (see also Table S1). (B) ChIP-seq signal of representative Bmal1 target genes in WT and Bmal1 knockout (Bmal1KO) liver. Original data were derived from published sources (Koike et al., 2012). (C) The frequency distribution of peak mRNA expression throughout the day of the 211 genes involved in mitochondrial function. (D) Diurnal expression of mitochondrial oxidative metabolism genes in control and LBmal1KO mice. Liver samples were collected every 4 hr for 24 hr (n=3-4/time point/genotype). The white and black bar represents light cycle and dark cycle, respectively. (E-F) Mitochondrial biogenesis in liver samples isolated at different time points determined by the relative Nd1 DNA content (E) or by flow cytometry analysis of MitoTracker Green staining. Data presented as mean ± SEM. *p<0.05.