Figure 1.
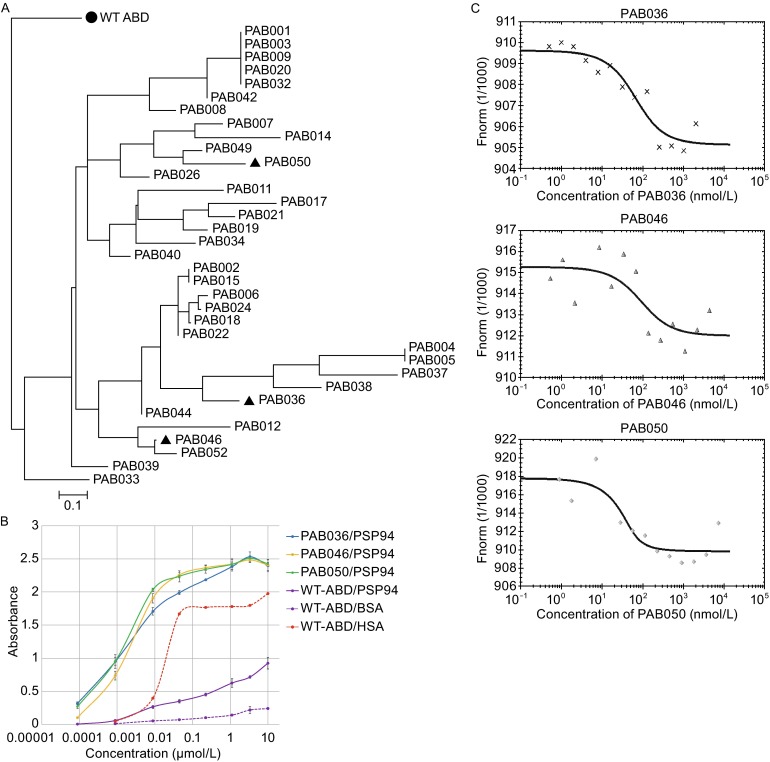
Generation of PAB variants and their binding to recombinant HisTag-PSP94. (A) Similarity tree of polypeptide sequences of the selected PAB binders. Analysis of a collection of 35 PAB binders obtained by ribosome display selection identified 29 unique sequence variants. For the analysis, randomized sequences between residues 20 and 46 were compared, as the N-terminal amino acid positions 1–19 were non-mutated. The sequence of the parental ABD wild-type domain (●) was used as a root of the tree. PAB variants selected for more detailed analysis are highlighted as triangles. (B) Binding of PAB variants to recombinant HisTag-PSP94 assessed by ELISA. Serially-diluted PAB variants in the form of biotinylated HisTag-PAB-TolA-AviTag fusion proteins were applied to a Polysorp microtiter plate coated with 10 μg/mL of recombinant HisTag-PSP94. WT-ABD indicates the parental wild-type ABD-TolA-AviTag protein as a non-mutated control with the natural affinity to HSA. The binding was detected by Streptavidin-HRP conjugate. The error bars represent the standard deviation from the three measurements. (C) The binding affinity of PAB variants to the fluorescently-labeled recombinant HisTag-PSP94 measured by microscale thermophoresis. Thermophoresis + T-jump data shown as binding curves were evaluated by a NanoTemper software and calculated Kd values for PAB036, PAB046, and PAB50 variants were 40 ± 6 nmol/L, 49 ± 10 nmol/L, and 10 ± 3 nmol/L, respectively
