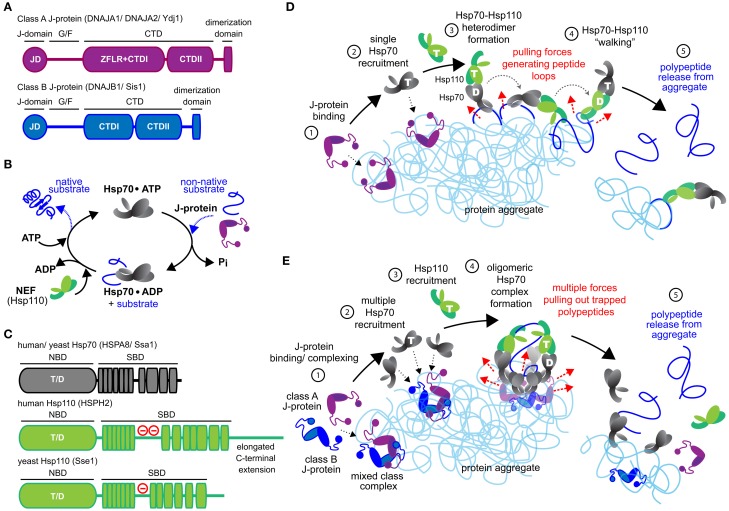
Figure 1.
Mechanistic models for Hsp70-based metazoan protein disaggregation. (A) Domain organization of class A and B J-proteins (as protomers). JD designates the conserved N-terminal J-domain. G/F denotes the glycine/phenylalanine rich flexible region; ZFLR, Zinc finger-like region; CTDI and CTDII, two homologous C-terminal β-sandwich substrate-binding domains. CTDs together with ZFLR provide substrate specificity. The dimerization domain forms functional J-protein homodimers. (B) Hsp70•J-protein•Hsp110 functional cycle. Concomitant interaction of Hsp70 with a J-protein and substrate results in allosteric stimulation of ATP hydrolysis trapping the substrate in Hsp70. Subsequent Hsp110 mediated ADP release from Hsp70 allows ATP rebinding, which triggers substrate release to complete Hsp70 cycle. (C) Schematic representation of the domain organization of yeast and human Hsp110 and Hsp70. (−) in red indicates the acidic region inserted between the terminal strands of the predicted β-sheet structure. The acidic loop determines the nuclear/cytoplasmic localization of human HSPH1 (Saito et al., 2009). The extended C-terminal domain is noted in HSPH2. ATP and ADP nucleotides bound to the NBD of Hsp70 and Hsp110s are denoted as “T” and “D,” respectively. (D) “Clamp and walk” model for Hsp70 and Hsp110 mediated protein disaggregation. Hsp70, J-protein and Hsp110 indicated in gray, purple and green, respectively. Nucleotide state at the NBDs of Hsp70 and Hsp110 indicated by T and D. Sequential reaction steps (encircled numbers): 1, J-protein targets aggregate; 2, J-protein recruits Hsp70; 3, Hsp110 recruitment and formation of Hsp70•Hsp110 heterodimer; 4, Hsp70•Hsp110 heterodimer “walking” on aggregate by alternating scanning (ATP state) and clamping (ADP state) substrate-interaction modes that generate pulling forces (dashed red arrows) on trapped polypeptides. Pulling forces result in forming peptide loops (dark blue) that fold to native-like conformations; 5, Releasing of polypeptides from aggregate due to accumulation of native-like folding events in trapped substrates. (E) Metazoan “nucleation” model for efficient Hsp70-based protein disaggregation. 1, J-protein targets and nucleates on aggregate; 2, Localized, multiple Hsp70 recruitment by J-protein assemblies on aggregates; 3, Hsp110 recruitment; 4, formation of oligomeric chaperone complex containing J-protein, Hsp110 and multiple Hsp70 molecules and buildup of entropic pulling forces (dashed red arrows) leading to extraction of trapped polypeptides (dark blue). 5, Hsp110 NEF activity triggered releasing of polypeptides from aggregate.